Subject Specific Sparse Dictionary Learning for Atlas Based Brain MRI Segmentation
Subject Specific Dictionary Learning for Atlas Based Brain MRI Segmentation
Snehashis Roy, Qing He, Elizabeth Sweeney, Aaron Carass, Daniel S. Reich, Jerry L. Prince, and Dzung L. Pham
Introduction
Quantitative measurements from segmentations of human brain magnetic resonance (MR) images provide important biomarkers for normal aging and disease progression. For example, quantitative measurements of brain tissues, such as gray matter, white matter, or the ventricles are important biomarkers in aging, dementia, and hypertension (S.M. Resnick et. al.) while white matter lesions and gray matter volumes are associated with the progression of Alzheimers disease and multiple sclerosis (N. Shiee et. al.). This is why segmentation of multiple tissues as well as lesions from MR images is important in research and potentially clinical settings. Segmentation is also important in many other image analysis procedures. In this paper, we propose a novel framework for dictionary-based multi-class segmentation of MR brain images. We call this method “Subject Specific Sparse Dictionary Learning” or “S3DL”. S3DL is an example-based approach, using patches as features and utilizing training data in the form of an MR image with a known segmentation.
Method
Unlike similar approaches (T. Tong et. al.) it employs dictionary learning to reduce the atlas size by selecting only the most relevant patches, leading to improved computational efficiency and accuracy. Also, S3DL can simultaneously segment multiple tissue classes while being informed by atlas priors which tell our algorithm where different anatomical structures are likely located within the image space.
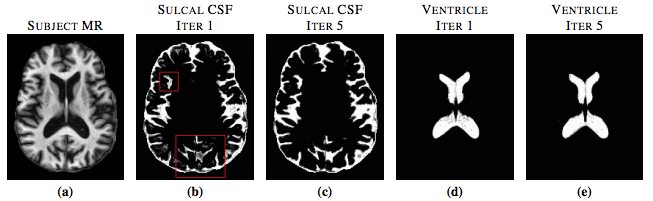
S3DL requires one set of MR images with a known segmentation to serve as training data, or in other words, an atlas. Then the atlas is then used in a machine learning framework to segment the subject image by matching patch-based features between the subject images and the atlas images. Because the atlas has a known anatomy, features in the atlas that are similar to a subject feature contribute information about the anatomy of the subject. We assume that for every subject patch a small number of similar looking patches can always be found from the collection of atlas patches (S. Roy et al.) (T. Cao et. al.). Sparse matching enforces the condition that every subject patch can be matched to only a few atlas patches. Previous methods have enforced similarity in spatial locations by searching for the similar patches in a small window around a specific voxel. We get around the need of this kind of windowed searching by including priors in the features. Our method enforces the similarity in texture between the subject patch and the chosen atlas patches. The previously described steps produce a segmentation that is influenced by the geometry of the spatial priors. However, because the priors are derived from a different brain image, it may not be ideally suited to the subject image because of the pathology or simply the variability of the brain geometry. So instead of using a fixed prior based on the initial atlas-to-subject registration, we dynamically update within an iterative loop. The priors at each iteration are replaced by a Gaussian blurred version of the obtained memberships, similar to the approach of (N. Shiee). The blurring relaxes the localization of the tissues in the memberships allowing for greater freedom in the segmentation computed at the next step. Figure 1 shows the effect of iteratively updating the priors via memberships.
These are the basic steps to the algorithm
1. At t = 0, we start with {a1, . . . , an+1, s1,![]() , . . . ,
, . . . ,![]() }, where
}, where ![]() are the registered atlas priors, k = 2, . . . , n + 1.
are the registered atlas priors, k = 2, . . . , n + 1.
2. Generate dictionaries ![]() and
and ![]() from Eqn. 5 using {a1, . . . , an+1, s1,
from Eqn. 5 using {a1, . . . , an+1, s1, ![]() , . . . ,
, . . . , ![]() }. The subject patches are denoted by b(0)(j).
}. The subject patches are denoted by b(0)(j).
3. At t ← t + 1, for each subject patch b(t)(j), generate the sparse coefficient x(t)(j) using ![]() from Eqn. 2.
from Eqn. 2.
4. Generate memberships {![]() , . . . ,
, . . . , ![]() } using x(t)(j)s from Eqn. 6
} using x(t)(j)s from Eqn. 6
5. Generate new adaptive priors ![]() ← Gσ
← Gσ ![]()
![]() , k = 2, . . . , n + 1. σ = 3mm is chosen empirically.
, k = 2, . . . , n + 1. σ = 3mm is chosen empirically.
6. Generate b(t+1)(j) using the updated priors {s1, ![]() , . . . ,
, . . . , ![]() }
}
7. Stop if ![]() , else go to step 3.
, else go to step 3.
Results
S3DL was implemented in Matlab (R2013a, The MathWorks, Natick, MA, USA) using parallel computation. For whole brain segmentation, the total runtime was typically 20 minutes on 2.7Ghz 12-core AMD processors for one 181 x 217 x 181 sized 1 mm3 image, of which approximately 10 minutes were spent on learning the dictionary. The run-time is reduced to 15 minutes for lesion segmentation for which dictionary learning takes approximately 5 minutes.
|
To determine the effect of dictionary size on the segmentation we varied it while segmenting Brainweb simulated images (C.A. Cocosco et. al.). If the dictionary size is too small the dictionary atoms may not represent the spectrum of the subject patches well. If too long, the computation time increases. We chose a Brainweb phantom with 0% noise and the same phantom with 3% noise or 5% noise was used to segment with varying dictionary sizes. We used various dictionary sizes from 1,000-6,000 and compared our results against the known segmentation. Around 5,000, the gained accuracy from increasing size plateaued so we then chose 5,000 as our dictionary size for the remainder of our experiments.
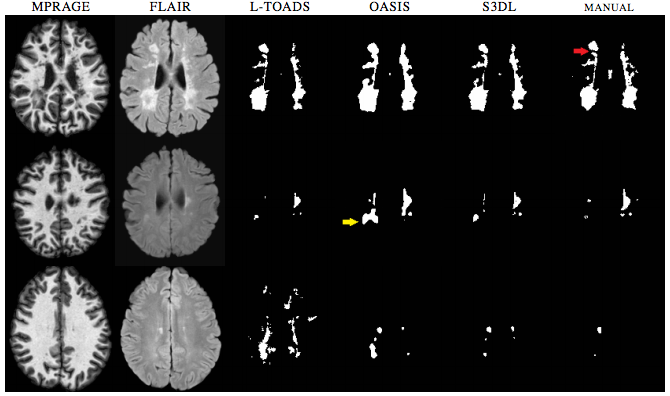
We then performed Lesion Segmentation with our algorithm. With this experiment we validated the dictionary learning aspect without priors on binary segmentation application, similar to (T. Tong et. al.). We compared the results of our lesion segmentations with LesionTOADS (N. Shiee et. al.) and OASIS (E.M. Sweeney et. al.). Examples of lesion segmentations are shown in Figure 2. While S3DL did make mistakes, when looking at the figures you can notice how it can be a balanced approach when compared to Oasis which while smooth can overestimate subtle changes in lesions and both OASIS and LesionTOADS show examples of gross over-estimation for which S3DL showed more visually accurate estimations.
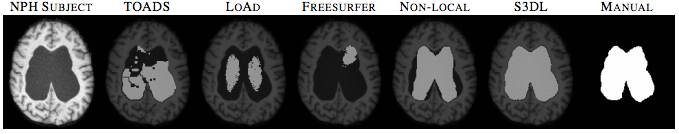
We also applied S3DL on 10 subjects with normal pressure hydrocephalus (NPH) having enlarged ventricles. Figure 3 shows one subject with five automated segmentations and the manual delineation of the ventricles. When you look at figure 10, you can see that visually, S3DL produces the most similar segmentation to the manual.
|
Conclusion
We have presented a patch-based sparse dictionary learning method for binary or multi-class MR brain image segmentation. The use of patches over single voxel intensities provides improved discrimination of anatomical structures. Contrary to previous patch-based segmentation methods, we use adaptive priors to localize different tissues with similar intensities as well as capture wide variabilities in anatomy between a subject and an atlas. We do not require any deformable registration of the subject atlas.
Software
Matlab executables are available.
Acknowledgments
Support for this work included funding from the Department of Defense in the Center for Neuroscience and Regenerative Medicine. This work is supported in part by the Intramural Research Program of the NIH and the grants NIH/NINDS R01NS070906, NIH/NIBIB R21EB012765, 1R01EB017743. We also thank Dr. Peter Calabresi for providing access the MS imaging data and Ms. Jennifer L. Cuzzocreo for help with lesion delineations.
References
- S.M. Resnick, D.L. Pham, M.A. Kraut, A.B. Zonderman, and C. Davatzikos. Longitudinal magnetic resonance imaging studies of older adults: A shrinking brain. Journal of Neuroscience, 23(8), 3295-3301, 2003.
- N. Shiee, P.L. Bazin, K.M. Zachowski, S.K. Farrell, D.M. Harrison, S.D. Newsome, J.N. Ratchford, B.S. Caffo, P.A. Calabresi, D.L. Pham, and D.S. Reich. Revisiting brain atrophy and its relationship to disability in multiple sclerosis. PtoS One. 7(5), e37049, 2012.
- T. Tong, R. Wolz, P. Coupe, J.V. Hajnal, D. Rueckert, and The Alzheimer's Disease Initiative. Segmentation of MR images via disciminative dictionary learning ad sparse coding: Applocation to hippocampus labeling. NeuroImage. 76(1), 11-23, 2013.
- S. Roy, A. Carass, and J.L. Prince. Magnetic resonance image example based contrast synthesis. IEEE Trans. Med. Imag. 32(12), 2348-2363, 2013.
- T. Cao, C. Zach, S. Modla, D. Powell, K. Czymmek, and M. Niethammer. Registration for correlative microscopy using image analogies. Workshop on biomedical image registration (WBIR). 296-306, 2012.
- N. Shiee, P.-L. Bazin, J. Cuzzocreo, A. Blitz, and D. Pham. Segmentation of brain images using adaptive atlases with application to ventriculomegaly. Inf. Pronc. in Medical Imaging. 1-12, 2011.
- C.A. Cocosco, V. Kollokian, R.K.S Kwan, and A.C. Evans. BrainWeb: Online interface to a 3D MRI simulated brain database. NeuroImage. 5(4), S425, 1997.
- N. Shiee, P. Bazin, A. Ozturk, D.S. Reich, P.A. Calabresi, and D.L. Pham. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 49(2), 1524-1535, 2009.
- E.M. Sweeney, R.T. Shinohara, N. Shiee, F.J. Mateen, A.A. Chudgar, J.L. Cuzzocreo, P.A. Calabresi, D.L. Pham, D.S. Reich, and C.M. Crainiceanu. OASIS is automated statistical inference for segmentation, with applications to multiple sclerosis. NeuroImage. 2, 402-413, 2013.
- P.L. Bazin and D.L. Pham. Homeomorphic brain image segmentation with topological and statistical atlases. Medical Image Analysis. 12(5), 616-625, 2008.
- M.J. Cardoso, M.J. Clarkson, G.R. Ridgway, M. Modat, N. C. Fox, and S. Ourselin. LoAd: A locally adaptive cortical segmentation algorithm. NeuroImage. 56(3), 1386-1397, 2011.
- A.M. Dale, B. Fischl, and M. I. Sereno. Cortical Surface-Based Analysis I: Segmentation and Surface Reconstruction. NeuroImage. 9(2), 179-194, 1999.
- P. Coupe, J.V. Manjon, J. Pruessner, M. Robles, V. Fonov, and D.L. Collins. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 54(2), 940-954, 2011.