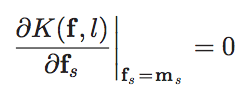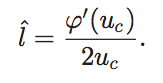Difference between revisions of "Reconstruction of high resolution tongue volumes from MRI"
| Line 12: | Line 12: | ||
Oral cancer may not have a high mortality rate, but the morbidity in terms of speech, mastication, and swallowing problems is significant and seriously affects the quality of life of the cancer patients. Characterizing the relationship between structure and function in the tongue is becoming a core requirement for both clinical diagnosis and scientific studies in the tongue/speech research community. A key to pursuing our goals is the ability to acquire a single high resolution 3D structural image of the tongue and vocal tract. A major problem with achieving a high resolution 3D structural image is time. A proper acquisition would take at least four to five minutes and after two to three minutes the tongue would start to involuntarily move, so therefore current protocol is to take three orthogonal volumes with axial sagittal and coronal orientations. But current protocol leads to no ideal image stack for 3D volumetric analyses. In particular, since each volume has poorer resolution in the slice-selection direction than in the in-plane direction, it is difficult to observe tongue muscles clearly in any one of the volumes. Therefore, reconstruction of a single high-resolution volumetric tongue MR image from the available orthogonal image stacks will improve our ability to visualize and analyze the tongue in living subjects. | Oral cancer may not have a high mortality rate, but the morbidity in terms of speech, mastication, and swallowing problems is significant and seriously affects the quality of life of the cancer patients. Characterizing the relationship between structure and function in the tongue is becoming a core requirement for both clinical diagnosis and scientific studies in the tongue/speech research community. A key to pursuing our goals is the ability to acquire a single high resolution 3D structural image of the tongue and vocal tract. A major problem with achieving a high resolution 3D structural image is time. A proper acquisition would take at least four to five minutes and after two to three minutes the tongue would start to involuntarily move, so therefore current protocol is to take three orthogonal volumes with axial sagittal and coronal orientations. But current protocol leads to no ideal image stack for 3D volumetric analyses. In particular, since each volume has poorer resolution in the slice-selection direction than in the in-plane direction, it is difficult to observe tongue muscles clearly in any one of the volumes. Therefore, reconstruction of a single high-resolution volumetric tongue MR image from the available orthogonal image stacks will improve our ability to visualize and analyze the tongue in living subjects. | ||
| − | + | ||
[[Image:SRT_Figure_1.png]] | [[Image:SRT_Figure_1.png]] | ||
''Figure 1'' | ''Figure 1'' | ||
| − | |||
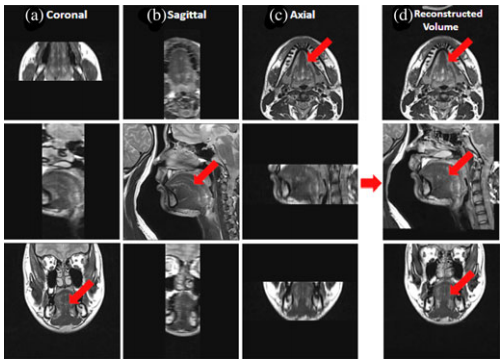
''Tongue images are acquired in three orthogonal volumes with FOV encompassing the tongue and surrounding structures. (a) Coronal, (b) sagittal, and (c) axial volumes are illustrated. The final superresolution volume using the proposed method is shown in (d). The red arrows indicate the tongue region.'' | ''Tongue images are acquired in three orthogonal volumes with FOV encompassing the tongue and surrounding structures. (a) Coronal, (b) sagittal, and (c) axial volumes are illustrated. The final superresolution volume using the proposed method is shown in (d). The red arrows indicate the tongue region.'' | ||
| − | + | ||
'''Method''' | '''Method''' | ||
| Line 29: | Line 28: | ||
The preprocessing steps are followed by a MAP-MRF reconstruction. Once three preprocessed, orthogonal volumes, regularized reconstruction is carried out. Classical methods including Tikhonov regularization have been used to achieve various image reconstruction objectives. But these methods often create oversmoothing of edges in direct opposition to our superresolution goals. Edge-preserving regularization methods, including total variation (TV), bilateral TV, and half quadratic regularization, have been developed to solve this problem. In this study, we used an edge-preserving regularization method of Villain et al., wherein MAP estimation with a half quadratic approach is used to obtain a high resolution volume. | The preprocessing steps are followed by a MAP-MRF reconstruction. Once three preprocessed, orthogonal volumes, regularized reconstruction is carried out. Classical methods including Tikhonov regularization have been used to achieve various image reconstruction objectives. But these methods often create oversmoothing of edges in direct opposition to our superresolution goals. Edge-preserving regularization methods, including total variation (TV), bilateral TV, and half quadratic regularization, have been developed to solve this problem. In this study, we used an edge-preserving regularization method of Villain et al., wherein MAP estimation with a half quadratic approach is used to obtain a high resolution volume. | ||
| − | + | ||
''Algorithm 1 MAP-MRF Reconstruction'' | ''Algorithm 1 MAP-MRF Reconstruction'' | ||
| Line 47: | Line 46: | ||
8. Iterate for loop until convergence | 8. Iterate for loop until convergence | ||
| − | + | ||
[[Image:SRT_Figure_4.png |frame|right|Figure 1]] | [[Image:SRT_Figure_4.png |frame|right|Figure 1]] | ||
| Line 65: | Line 64: | ||
| − | + | ||
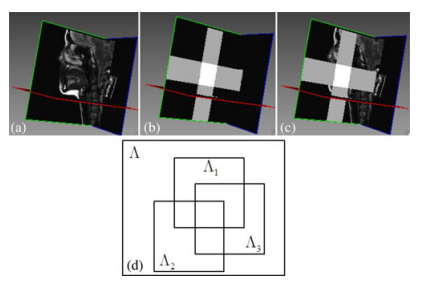
''Fig. 2: (a) One representative final superresolution image; (b) regions defined from each low-resolution volume; (c) volume with regions defined from each low resolution volume; and (d) schematic drawing of each region.'' | ''Fig. 2: (a) One representative final superresolution image; (b) regions defined from each low-resolution volume; (c) volume with regions defined from each low resolution volume; and (d) schematic drawing of each region.'' | ||
| − | + | ||
In order to gather our tongue data in this study, fifteen high-resolution MR datasets were used. They came from twelve normal speakers and three patients who had tongue cancer surgically resected. The FOV was 240mm x 240mm with a resolution of 256x256. The proposed method was first evaluated using fifteen simulated datasets. In order to quantitatively evaluate the performance of the proposed method in terms of accuracy of the reconstruction, the final superresolution volume using the proposed method was considered as our ground truth data. The ground truth was constructed with 256x256x256 voxels with a resolution of 0.94mm x 0.94mm x 0.94mm. In what follows, volume reconstruction was carried out in four ways. first, fifth-order B-spline interpolation was performed in each plane independently. Second, averaging of three up-sampled volumes was performed. Third, reconstruction from three up-sampled volumes using Tokhonov regularization was performed. Finally, the proposed method using three upsampled volumes was carried out. | In order to gather our tongue data in this study, fifteen high-resolution MR datasets were used. They came from twelve normal speakers and three patients who had tongue cancer surgically resected. The FOV was 240mm x 240mm with a resolution of 256x256. The proposed method was first evaluated using fifteen simulated datasets. In order to quantitatively evaluate the performance of the proposed method in terms of accuracy of the reconstruction, the final superresolution volume using the proposed method was considered as our ground truth data. The ground truth was constructed with 256x256x256 voxels with a resolution of 0.94mm x 0.94mm x 0.94mm. In what follows, volume reconstruction was carried out in four ways. first, fifth-order B-spline interpolation was performed in each plane independently. Second, averaging of three up-sampled volumes was performed. Third, reconstruction from three up-sampled volumes using Tokhonov regularization was performed. Finally, the proposed method using three upsampled volumes was carried out. | ||
| Line 78: | Line 77: | ||
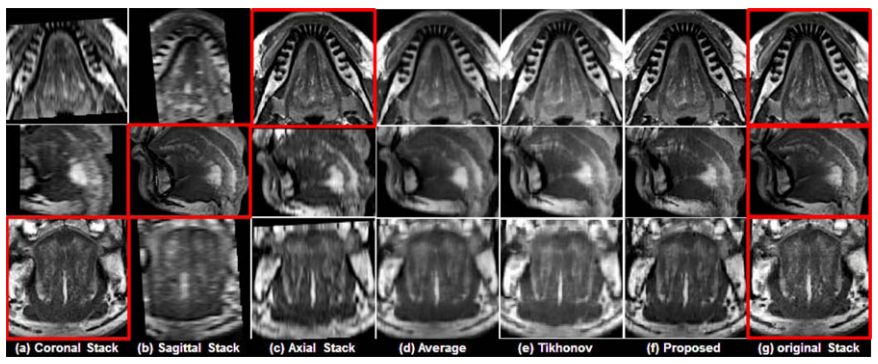
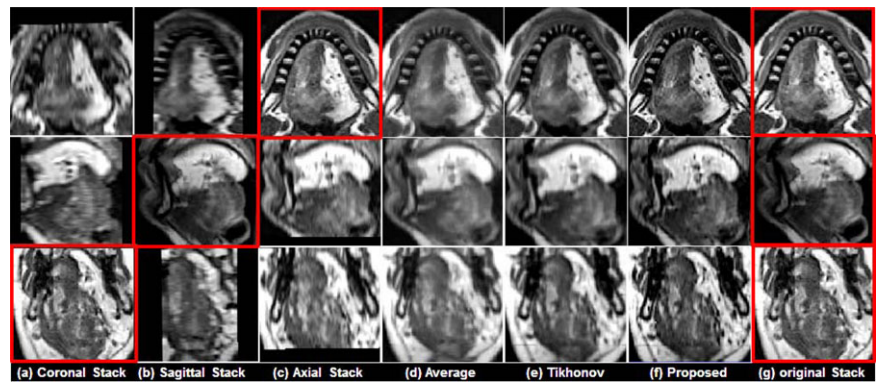
See figs. 3 and 4, two representative results using a normal subject (see fig. 3) and a glossectomy patient (see fig. 4) with different reconstruction methods demonstrated, respectively. The rows show slices of three orthogonal views including axial , sagittal, and coronal, respectively. The first three columns (a)-(c) show three original scans after isotropic resampling in the coronal, sagittal, and axial planes, respectively. Panel (d) shows the reconstruction using averaging, (e) shows reconstruction using Tikhonov regularization and (f) shows reconstruction using the proposed method. Panal (g) hows the original orthogonal volumes. | See figs. 3 and 4, two representative results using a normal subject (see fig. 3) and a glossectomy patient (see fig. 4) with different reconstruction methods demonstrated, respectively. The rows show slices of three orthogonal views including axial , sagittal, and coronal, respectively. The first three columns (a)-(c) show three original scans after isotropic resampling in the coronal, sagittal, and axial planes, respectively. Panel (d) shows the reconstruction using averaging, (e) shows reconstruction using Tikhonov regularization and (f) shows reconstruction using the proposed method. Panal (g) hows the original orthogonal volumes. | ||
| − | + | ||
[[Image:SRT_Figure_5.png]] | [[Image:SRT_Figure_5.png]] | ||
''Figure 3'' | ''Figure 3'' | ||
| − | + | ||
''Comparison of different reconstruction methods on a normal subject. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods including simple averaging, Tikhonov regularization, and the proposed method are shown in (d), (e), and (f), respectively. For reference, the highest resolution images from each original volume are repeated in (g).'' | ''Comparison of different reconstruction methods on a normal subject. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods including simple averaging, Tikhonov regularization, and the proposed method are shown in (d), (e), and (f), respectively. For reference, the highest resolution images from each original volume are repeated in (g).'' | ||
| − | + | ||
[[Image:SRT_Figure_6.png]] | [[Image:SRT_Figure_6.png]] | ||
''Figure 4'' | ''Figure 4'' | ||
| − | + | ||
''Comparison of different reconstruction methods using a glossectomy patient. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods include (d) simple averaging,(e) Tikhonov regularization, and (f) the proposed method, respectively. For reference, the highest resolution images from each original volume are repeated in (g).'' | ''Comparison of different reconstruction methods using a glossectomy patient. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods include (d) simple averaging,(e) Tikhonov regularization, and (f) the proposed method, respectively. For reference, the highest resolution images from each original volume are repeated in (g).'' | ||
| − | + | ||
| + | |||
The proposed method provided better muscle and fine anatomical detail than the other methods. The target reference volume into which the other volumes were registered was the axial volume in both cases. In our experiments, preprocessing steps (e.g., registration) were not validated in a quantitative manner. Instead, the quality of the results was confirmed visually. Notice that the reconstructed images and the original mages shown in the figures may not be exactly the same except in the axial slice, because the data are aligned to the axial stack. In our experimental results, patients have more complex anatomy than controls. They have considerable scar tissue, their muscles are deformed around the scar and missing in the area of resection, and they are asymmetrical. Therefore, it was possible that their datasets would have been more difficult to align. But it is seen from our experimental results that all were reconstructed with equally good success. | The proposed method provided better muscle and fine anatomical detail than the other methods. The target reference volume into which the other volumes were registered was the axial volume in both cases. In our experiments, preprocessing steps (e.g., registration) were not validated in a quantitative manner. Instead, the quality of the results was confirmed visually. Notice that the reconstructed images and the original mages shown in the figures may not be exactly the same except in the axial slice, because the data are aligned to the axial stack. In our experimental results, patients have more complex anatomy than controls. They have considerable scar tissue, their muscles are deformed around the scar and missing in the area of resection, and they are asymmetrical. Therefore, it was possible that their datasets would have been more difficult to align. But it is seen from our experimental results that all were reconstructed with equally good success. | ||
Revision as of 19:18, 4 June 2014
<meta name="title" content="Reconstruction of High-Resolution Tongue Volumes From MRI"/>
Reconstruction of High-Resolution Tongue Volumes From MRI
Jonghye Woo, Emi Z Murano, Maureen Stone, and Jerry L Prince
This page is under construction.
Introduction
Oral cancer may not have a high mortality rate, but the morbidity in terms of speech, mastication, and swallowing problems is significant and seriously affects the quality of life of the cancer patients. Characterizing the relationship between structure and function in the tongue is becoming a core requirement for both clinical diagnosis and scientific studies in the tongue/speech research community. A key to pursuing our goals is the ability to acquire a single high resolution 3D structural image of the tongue and vocal tract. A major problem with achieving a high resolution 3D structural image is time. A proper acquisition would take at least four to five minutes and after two to three minutes the tongue would start to involuntarily move, so therefore current protocol is to take three orthogonal volumes with axial sagittal and coronal orientations. But current protocol leads to no ideal image stack for 3D volumetric analyses. In particular, since each volume has poorer resolution in the slice-selection direction than in the in-plane direction, it is difficult to observe tongue muscles clearly in any one of the volumes. Therefore, reconstruction of a single high-resolution volumetric tongue MR image from the available orthogonal image stacks will improve our ability to visualize and analyze the tongue in living subjects.
Figure 1
Tongue images are acquired in three orthogonal volumes with FOV encompassing the tongue and surrounding structures. (a) Coronal, (b) sagittal, and (c) axial volumes are illustrated. The final superresolution volume using the proposed method is shown in (d). The red arrows indicate the tongue region.
Method
In this study we develop a fully automated and accurate superresolution volume reconstruction method from three orthogonal image stacks of the same subject by extending our preliminary approach. We present a refined preprocessing and reconstruction algorithm design and provide validations on both tongue and brain data. We use a number of preprocessing steps including motion correction, intensity normalization, etc., followed by a region based maximum a posteriori (MAP) Markov random field (MRF) approach. The region based approach allows us to reconstruct the tongue at high resolution, because it resides in the intersection of the image FOVs, as well as other regions around the tongue which are used in the analysis of speech production. In order to preserve important anatomical features such as subtle boundaries of muscle, we use edge preserving regularization. To our knowledge, this is the first attempt at superresolution reconstruction applied to in vivo tongue high-resolution MR images for both normal and diseased subjects. The resulting superresolution volume improves both the signal-to-noise ratio (SNR) and resolution over the source images, thereby approximating an original high-resolution volume whose acquisition would have taken too long for the subject to refrain from swallowing (or making other motions).
In the preprocessing procedure we go through five steps prior to the reconstruction of the superresolution volume. First is a generation of an isotropic volume followed by conversion of the orientation of each volume to the orientation of the target reference volume. Then, padding of zero values to yield the same volume sizes and afterwords registration to correct subject morion between volumes. Finally, matching of the intensities in the overlap region using spline regression is done.
The preprocessing steps are followed by a MAP-MRF reconstruction. Once three preprocessed, orthogonal volumes, regularized reconstruction is carried out. Classical methods including Tikhonov regularization have been used to achieve various image reconstruction objectives. But these methods often create oversmoothing of edges in direct opposition to our superresolution goals. Edge-preserving regularization methods, including total variation (TV), bilateral TV, and half quadratic regularization, have been developed to solve this problem. In this study, we used an edge-preserving regularization method of Villain et al., wherein MAP estimation with a half quadratic approach is used to obtain a high resolution volume.
Algorithm 1 MAP-MRF Reconstruction
1. Initialization
2. For each pixel s on the volume do
3._ Determine overlapping volumes using regions defined in Fig. 2
4._ Calculate ![]() according to Eq. 1
according to Eq. 1
5._ Update ![]() according to the over-relaxation scheme in Eq. 2
according to the over-relaxation scheme in Eq. 2
6._ For every clique ![]() , including
, including ![]() , update
, update ![]() according to Eq. 3
according to Eq. 3
7. end for
8. Iterate for loop until convergence
Fig. 2: (a) One representative final superresolution image; (b) regions defined from each low-resolution volume; (c) volume with regions defined from each low resolution volume; and (d) schematic drawing of each region.
In order to gather our tongue data in this study, fifteen high-resolution MR datasets were used. They came from twelve normal speakers and three patients who had tongue cancer surgically resected. The FOV was 240mm x 240mm with a resolution of 256x256. The proposed method was first evaluated using fifteen simulated datasets. In order to quantitatively evaluate the performance of the proposed method in terms of accuracy of the reconstruction, the final superresolution volume using the proposed method was considered as our ground truth data. The ground truth was constructed with 256x256x256 voxels with a resolution of 0.94mm x 0.94mm x 0.94mm. In what follows, volume reconstruction was carried out in four ways. first, fifth-order B-spline interpolation was performed in each plane independently. Second, averaging of three up-sampled volumes was performed. Third, reconstruction from three up-sampled volumes using Tokhonov regularization was performed. Finally, the proposed method using three upsampled volumes was carried out.
Brain Data was also collected. Three isotropic high-resolution brain MR datasets were used to objectively evaluate and compare the performance of different reconstruction methods. For each subject, two magnetization prepared rapid gradient echo (MPRAGE) images were obtained. One challenge in the proposed method is a lack of ground truth available in in vivo volumetric tongue MR data. In order to avoid potential bias in our experiments, we used two high-resolution brain datasets with isotropic resolution to evaluate the performance of different reconstruction methods.
Results
See figs. 3 and 4, two representative results using a normal subject (see fig. 3) and a glossectomy patient (see fig. 4) with different reconstruction methods demonstrated, respectively. The rows show slices of three orthogonal views including axial , sagittal, and coronal, respectively. The first three columns (a)-(c) show three original scans after isotropic resampling in the coronal, sagittal, and axial planes, respectively. Panel (d) shows the reconstruction using averaging, (e) shows reconstruction using Tikhonov regularization and (f) shows reconstruction using the proposed method. Panal (g) hows the original orthogonal volumes.
Figure 3
Comparison of different reconstruction methods on a normal subject. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods including simple averaging, Tikhonov regularization, and the proposed method are shown in (d), (e), and (f), respectively. For reference, the highest resolution images from each original volume are repeated in (g).
Figure 4
Comparison of different reconstruction methods using a glossectomy patient. The original coronal, sagittal, and axial volumes are shown in (a), (b), and (c), respectively. B-spline interpolation was used to yield higher pixel resolutions equal to that of the highest resolution orientations, which are indicated by the red boxes. Three different reconstruction methods include (d) simple averaging,(e) Tikhonov regularization, and (f) the proposed method, respectively. For reference, the highest resolution images from each original volume are repeated in (g).
The proposed method provided better muscle and fine anatomical detail than the other methods. The target reference volume into which the other volumes were registered was the axial volume in both cases. In our experiments, preprocessing steps (e.g., registration) were not validated in a quantitative manner. Instead, the quality of the results was confirmed visually. Notice that the reconstructed images and the original mages shown in the figures may not be exactly the same except in the axial slice, because the data are aligned to the axial stack. In our experimental results, patients have more complex anatomy than controls. They have considerable scar tissue, their muscles are deformed around the scar and missing in the area of resection, and they are asymmetrical. Therefore, it was possible that their datasets would have been more difficult to align. But it is seen from our experimental results that all were reconstructed with equally good success.
Conclusion
In this study, a superresolution reconstruction technique based on the region based MAP-MRF with an edge-preserving regularization and half quadratic approach was developed for volumetric tongue MR images acquired from three orthogonal acquisitions. Experimental results show that the proposed method has superior performance to the interpolation-based method, simple averaging, and Tikhonov regularization. It It also better preserved the anatomical details as quantitatively confirmed. The proposed method allows full 3-D high resolution volumetric data, thereby potentially improving further image/motion and visual analyses.
Acknowledgements
The authors would like to thank S. Ying, Department of Radiology, Johns Hopkins University, for sharing brain data in their simulation. They would also like to thank the reviewers for their helpful comments
References
- Jonghye Woo et al., “Reconstruction of High-Resolution Tongue Volumes From MRI” IEEE Transactions On Biomedical Engineering., 59, no. 12 (2012). 3511-3524
- N. Villain et al., “Three-dimensional edge preserving image enhancement for computed tomography,” IEEE Trans. Med. Imag. 22, no.10 (2003). 1275-1287.
- M Stone et al., “A preliminary application of principal components and cluster analysis to internal tongue deformation patterns,” Comput. Methods Biomech. Biomed. Eng., 13, no 4 (2010). 493-503.
- N. Nguyen et al., “A computationally efficient super-resolution image reconstruction algorithm” IEEE Trans. Imag. Process., 10, no.4 (2001). 573-583.