
 CATNAP : Screen Shots
CATNAP : Screen Shots
(Coregistration,
Adjustment, and Tensor-solving, a Nicely Automated Program)
[Home] [Back/Research] [Back/Catnap]
CATNAP FAQ/ QAU
(Frequently Asked Questions and Questions Asked Once)
Revision History:
8/14/2006 Initial
Document / BL
8/15/2006 updated / JF
12/11/2006 updated /
BL
Q: What is CATNAP and what does it do?
CATNAP was developed to meet the needs of DTI researchers. CATNAP provides three principal services. 1) image coregistration, 2) DTI gradient table creation and correction 3) Diffusion tensor calculations. By bundling these steps and linking them with a GUI, CATNAP provides a quick and efficient way to process DTI data and coregister anatomical MR scans.
Q: Can I run CATNAP from the command line?
Yes, all CATNAP functionality is available from the command line and can be implemented in batch and script processing.
Q. What do all the file extensions correspond to
in CATNAP?
Acquired Data Files (.par, .rec)
The acquired data from the Philips scanner is in .par and .rec format. The .par file is in ASCII text format and contains useful information about the scan protocol. The .rec file is in int16 format and contains the MR data.
Intermediate Data Files (.img, .hdr, .frec, .parv2)
CATNAP splits the .rec data into analyze format files (.img and .hdr). These files are created and then subsequently removed once motion correction is complete. .frec files are the MR data in floating point format. The floating point data is generated by scaling the .rec file by the parameters listed in the .par file. Coregistration is performed with the .frec files. The .parv2 file is a custom built file that mirrors the .par file in content, but is easier to parse. Anything with RDR1 prefix denotes a file that has been coregistered.
Final Data Files (.xfm, _sc1.rec, _sc1.par, .frec, .parv2)
The transformation matrix recorded from FSL FLIRT is saved as a .xfm file. This file is needed to correct the gradient tables. The coregistered DTI data is scaled (dived by 10) to fit within the constraints of int16 format.
Q: Should I read in the RDR1*_sc1.rec file with
readrec or readraw?
As the _sc1.rec file has already been scaled by the values in the par file, you should use readraw to read in the _sc1.rec file. The _sc1.par file is provided as a reference for image matrix size and FOV parameters.
Q: What do I do if I want report a problem?
Please direct your email correspondence to Bennett Landman or Jonathan Farrell. Please include as much information about your problem as possible, including the path to the original data (if using Godzilla), screen capture of problem (if applicable), and the steps you implement to reproduce the error.
Q: Do I need to cite CANTAP and how do I cite
CATNAP?
The developers would appreciate an acknowledgement of their many hours of work and continued development time on the CATNAP project. If CATNAP has been useful to your project, saved you time, limited your gray hair, alleviated heartburn and otherwise made DTI processing simple, please cite CATNAP as follows.
Acknowledgement: The
CATNAP software was kindly provided by Bennett Landman et al, (
Citation: B. A. Landman, J. A. Farrell, C. K. Jones, S. A. Smith, J. L. Prince, P. C. van Zijl, and S. Mori. “Effects of Diffusion Weighting Schemes on the Reproducibility of DTI-derived Fractional Anisotropy, Mean Diffusivity, and Principal Eigenvector Measurements at 1.5T”, NeuroImage. 36(4): 1123-1138. July 2007.
In text reference (Landman et al, 2007)
Q: Does CATNAP
support coronal or sagittal par/rec files?
A: No. We are looking into this. As of CATNAP 2.1, only axial DTI and axial anatomical datasets are supported for image coregistration. Future releases will likely support automatic slice orientation detection. If you have sagital or coronal DTI data, you can utilize Jonathan Farrell’s DTI_gradient_table_creator.m code (in matlab) to construct the correct gradient tables.
Q: How does CATNAP resample/reslice the data
being coregistered?
When CATNAP runs FSL FLIRT, any anatomical scans that are coregistered to the DTI data will be output to have the same resolution as the DTI data (same in plane resolution, same number of voxels and same slice thickness). Therefore an MPRAGE dataset with 1mm slices will be resampled to 2mm slice thicknesss if the DTI dataset has 2mm slices. This ensures that there is a one to one correspondence of voxels in coregistered data, which is very useful for ROI placement and fiber tracking applications.
Q: How do I skip the
registration step?
A: In order for the gradient table adjuster to locate all necessary files, we need to have the file nomenclature and directory structure identical to that which would have been created if you press the “Perform Motion Correction” button. So, to use already registered files, you must select the files that are not yet registered (typically the .par files) and make sure that the properly named registered ones already exist (typically the .parv2 and .frec files). We realize that this may be counter intuitive, but this is the most robust way be can think of to force proper nomenclature. Once a par/rec file has been registered to a particular output directory with this system, additional analyses should be able to be performed without too much hassle. With a bit of careful naming and storage of transform files, external registration schemes maybe used as well.
Q: I am unable to
load files and get an error as attached: Undefined command/function 'loadPAR'?
A:
This looks like it cannot find the loadPAR.m file. Did you unzip the 4 folders?
(You need them all.) CATNAP uses modular toolboxes to load/save files, use
Analyze images, and provide general functions. All these mini-toolboxes need to
be added to your Matlab path. I typically use: addpath(genpath('ROOT')); Where
ROOT is the path containing the four folders. The folders are:
BennettUtility – My handy utility
functions
fileformats_toolbox – load/save
various data formats
MRI – Darren Webber’s MRI_TOOLBOX
(slightly modified)
Q: There is also an
error message that says a vertical resolution of at least 1024 is required?
A: Matlab does not robustly support resizing of graphical user interfaces. On current versions of Matlab, if a figure is too large for the screen, it is resized and the components become unusable. CATNAP is 710x855, so we display a warning message if your reported screen resolution is less than 1024 lines vertically (to leave room for toolbars, etc). However, we still attempt to let Matlab do the right thing and CATNAP may or may not work if you ignore this message. A final rub is that on some systems, Matlab does not accurately report the screen resolution, so you will see this message even though there are no resizing problems.
Q: The DTI
processing must have flipped a sign somewhere, because the color maps are all
wrong. Not sure where this could have been, or perhaps I didn't set things up
properly for that phase. What could be going wrong?
A:
The DTI color directions are determined by the parameters that you specify in
the "DTI Adjustment" section.
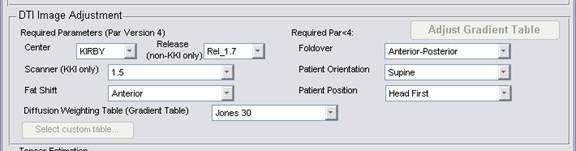
Perhaps you have an incorrect direction: fat shift, foldover, patient orientation, or patient position. (You need to check the LIST files that the scanner dumps. These data are not in the PAR files.) If you are certain of your choices, please send them to Jon Farrell to make sure that we correctly process your case.
Q: I register DTI scans using Philips Pride
software ( format is .rec for the registered scans) and I would like to further
analyse this with CATNAP. I do not have the .frec and .parv2 file format
for my registered file.How should I proceed?
A: If you use an outside registration algorithm, then you need to also export the transforms that were used to rotate the original scan. A principle complexity in registering DTI images is that you also need to correct the diffusion weighting directions when you transform a diffusion weighted file.
You may use any 3rd party registration file as long as you:
1) export the image data into par/rec
2) save the transform files as “RDR_<par/rec name>_VolXXX.xfm” in the “xfm” subdirectory of your target directory. There must be one xfm file (text file, 4 rows, space delimited coefficients of the affine transform)
3) check the “selected files are motion corrected” checkbox in CATNAP
4) save the par/rec files as par/frec files with a “RDR_” prefix in the target directory pr = loadPARREC(<filename>); savePARfREC(pr,[TARGET_DIR filesep ‘RDR_’ <filename’]);
In order to see the exact filenames and structures required, it may be useful to run CATNAP without the “selected files are motion corrected” checkbox.
Q: Does CATNAP work
with Siemens or GE dicom images?
A: We do not have specific expertise using diffusion weighting tables from non-Philips scanners, so you will need to be able to identify your diffusion weighting schemes relative to the slice layout. From there, using CATNAP is not difficult.
At least one group is successfully using CATNAP with Siemens DICOM files. To do so, you need to convert your data to Analyze (for example, with MIPAV from the NIH: http://mipav.cit.nih.gov/). Then use Analyze2PARREC in the CATNAP distribution to convert to PARREC (the Philips research format). To use the CATNAP gradient table correction system, click “Table is relative to slice.”
Q: Some of the contrasts appear blank when loaded in DTIStudio even
when the level is adjusted. I can see the data if I load the contrast in
Matlab. What is going on?
A: This is a “feature” of DTIStudio. When you read in any contrast in DTIStudio, it automatically quantizes it. This means that is finds the max and min and them picks a set number of values in between (254, I believe) and rounds all data to these values. If the range is REALLY big, then all meaningful values may be rounded to zero, so there may be no way of adjusting the scaling and see the data.
To get around this problem, you can: pre-clip the data to remove the effects of noise from outside the head:
function clipData(filenameIn, filenameOut)
% Load the data
Dat = loadFloat(filenameIn); % to use my toolbox, load the data anyway you like.,
% Find the Quantiles
Temp = sort(Dat(:));
% Clip the data to between the 5th and 95th quantile – you can replace these with whatever you want.
% for example, the absolute values 0 to 1 make sense for FA
Dat = min(max(Dat, Temp(floor(.05*length(Temp)))), Temp(floor(0.95*length(Temp))));
% Write out the data
fp = fopen(filenameOut, 'wb'); fwrite(fp,Dat(:),'float');
Q: In our case, if we use a high resolution anatomical scan as our
coregistration "target" and DTI scans as sources, does that mean
after this step, DTI scans are modified so that it was aligned to the
anatomical?
A: Short Answer: Yes.
If:
DTI is 128x128x32x30 voxels sized 2.2x2.2x2.2 mm Structural is 256x256x70 sized 1.1x1.1x1 mm
Case 1: Target "First B0"
Result:
DTI is 128x128x32x30 sized 2.2x2.2x2.2 mm Structural is 128x128x32 sized 2.2x2.2x2.2 mm
Case 2: Target is "Other Volume" with the structural file selected
Result:
DTI is 256x256x70x30 sized 1.1x1.1x1 mm
Structural is 256x256x70 sized 1.1x1.1x1 mm
Long Answer: All the above is true, except...
The real way that we get the transforms is to register to target to the structural and the invert the transform. We find this is more stable when the target tends to have the same or more FOV coverage than the source. Only the SRCs (files in the DTI and structural boxes) in target space are ever written out.
Q: I tried running CATNAP in the linux system. I was able to perform
motion correction and i got a message saying "motion correction
complete". I thus see an xfm folder created. However, i do not see a
scaled and dti folder. I get the
following error message after motion correction: "Please enter:
yes-ovp-high, yes-ovp-medium, yes-ovp-low..........or user defined for first
inoput"
A: The “jones30” scheme is not available by default on “OTHER” scanners (meaning outside of KKI). If you used Jones30 in user defined mode, then you must specify the table as user defined. Please choose the diffusion weighting scheme that you used.
Last Updated: Tuesday May 27, 2008
© Copyright 2007, Bennett Landman. All rights reserved.