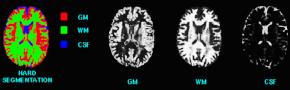
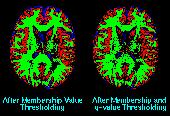
Statistical Characterization of Brain Tissues in Magnetic Resonance Imaging
D.L. Pham, J.L. Prince, A.P. Dagher, and C. Xu
|
Overview
|
||||||||||||
|
||||||||||||
|
Introduction
|
||||||||||||
| Magnetic resonance
imaging (MRI) is a highly successful diagnostic imaging modality,
largely due to its ability to derive contrast from a number
of physical parameters. An understanding of how these parameters
vary with tissue type, pathology, and other factors is therefore
fundamental to determining how MRI can best be used for diagnostic
purposes.
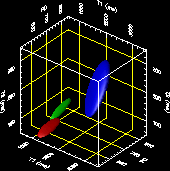
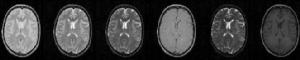
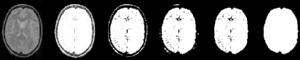
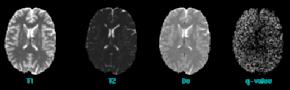
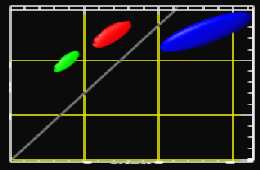
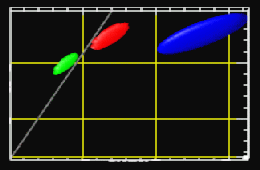
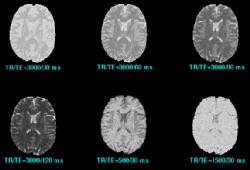
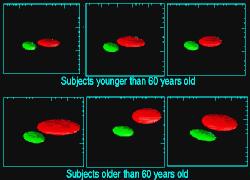
In MRI, tissue contrast is based predominantly on the spin-lattice relaxation time (T1), spin-spin relaxation time (T2), and proton spin density (PD) of the tissues being imaged. Differences in these parameters, particularly in brain tissues, have been correlated with differences in age, sex, and disease. In addition, statistics on these parameters have been used in the optimization of pulse sequences for improved image contrast and segmentation. Further research is required on the variability of tissue parameters across populations and the evolution of parameters with pathology and age. Such research would provide invaluable information on not only biochemical processes and physiology, but also on how MR images can be interpreted and improved. We describe a procedure for automatically estimating the conditional joint pdf of T1, T2, and PD, given that the tissue is either GM, WM, or CSF in the brain. A large number of tissue parameter estimates are accumulated using a multi-slice, multi-echo MRI acquisition. Two criteria are introduced for disregarding data which have been corrupted by partial volume averaging, noise, or other artifacts. The procedure is fully automated, making studies involving a large number of subjects feasible. |
||||||||||||
|
Procedure
|
||||||||||||
|
||||||||||||
|
Applications
|
||||||||||||
|
||||||||||||
|
Conclusion
|
||||||||||||
| A procedure for characterizing the NMR tissue parameters of GM, WM, and CSF in the brain as a conditional joint pdf has been described. The procedure is completely automated and compensates for MRI artifacts by using two thresholding criteria. Example applications were shown employing the joint pdf's for studying the variability of tissue parameters across subjects, pulse sequence optimization, and computational phantoms. Additional experiments, including phantom studies, are necessary to establish the precision and accuracy of the procedure. A comprehensive longitudinal study across a large number of patients applying the procedure could prove highly revealing with respect to the variability of tissue parameters in the brain. | ||||||||||||
| Publications | ||||||||||||
|