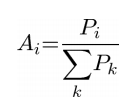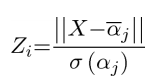Difference between revisions of "Principal Component Analysis of Cerebellar Shape"
| (7 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | <meta name="title" content="Principal Component Analysis of Cerebellar Shape"/> | + | <!-- <meta name="title" content="Principal Component Analysis of Cerebellar Shape"/> --> |
{{h2|Principal Component Analysis of Cerebellar Shape}} | {{h2|Principal Component Analysis of Cerebellar Shape}} | ||
| Line 5: | Line 5: | ||
{{TOCright}} | {{TOCright}} | ||
| − | |||
| − | |||
{{h3|Introduction}} | {{h3|Introduction}} | ||
| Line 21: | Line 19: | ||
Regions of interest were manually delineated using Display software by two raters who were blinded to the diagnosis of participants. Inter-rater reliability of the volumetric measurements was determined by having each rater analyze four subjects for determination of intraclass correlation coefficients. Regions of interest were visually identified based on Schmahmann’s cerebellar atlas. The total cerebellar volume was defined to include the cerebellar cortex, arbor vitae corpus medullare, and deep cerebellar nuclei. In the presence of cerebellar atrophy, the volumetric measurements excluded the cerebrospinal fluid space between the lobules of the cerebellum. | Regions of interest were manually delineated using Display software by two raters who were blinded to the diagnosis of participants. Inter-rater reliability of the volumetric measurements was determined by having each rater analyze four subjects for determination of intraclass correlation coefficients. Regions of interest were visually identified based on Schmahmann’s cerebellar atlas. The total cerebellar volume was defined to include the cerebellar cortex, arbor vitae corpus medullare, and deep cerebellar nuclei. In the presence of cerebellar atrophy, the volumetric measurements excluded the cerebrospinal fluid space between the lobules of the cerebellum. | ||
| − | |||
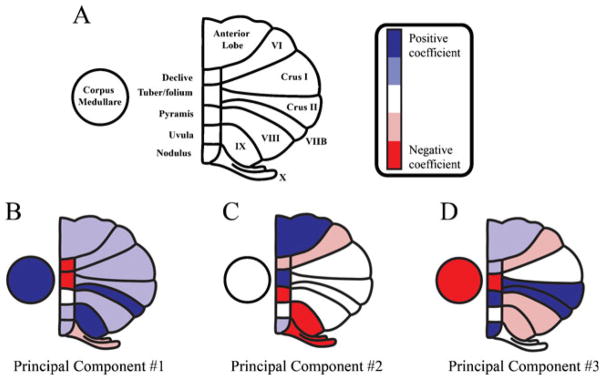
| − | '''Figure 1''': Principal component coefficients of the first three principal components. a An illustration of cerebellum with labeled subregions. Colormap indicates principal component coefficients or loadings. (The coefficient of each cerebellar subregion was classified into five equal-size intervals from minimum (coefficients) to maximum (coefficients) for each principal component [dark blue: positive coefficients (highest coefficient value); red: negative coefficients (lowest coefficient value)].) Principal component coefficients represent the relative “weight” of each original variable (relative cerebellar regional volume) in each principal component. The principal component coefficients are shown for the first three principal components (b–d) | + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> |
| + | {| | ||
| + | |- | ||
| + | | align=center|[[Image:PCA_Figure_2.jpg]] | ||
| + | |- | ||
| + | |'''Figure 1''': Principal component coefficients of the first three principal components. a An illustration of cerebellum with labeled subregions. Colormap indicates principal component coefficients or loadings. (The coefficient of each cerebellar subregion was classified into five equal-size intervals from minimum (coefficients) to maximum (coefficients) for each principal component [dark blue: positive coefficients (highest coefficient value); red: negative coefficients (lowest coefficient value)].) Principal component coefficients represent the relative “weight” of each original variable (relative cerebellar regional volume) in each principal component. The principal component coefficients are shown for the first three principal components (b–d) | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| Line 31: | Line 36: | ||
To validate the hypothesis that PCA can correctly classify disease groups based on cerebellar shape characteristics alone, we performed a bootstrap validation, leaving out each subject ([[Image:PCA_Subjectbootstrap.png]]) one at a time. The PCA space was defined without the anatomic information of [[Image:PCA_Subjectbootstrap.png]]. The relative regional volumes of [[Image:PCA_Subjectbootstrap.png]] were regressed against the anatomic scores of all subjects minus [[Image:PCA_Subjectbootstrap.png]] within the PCA space to calculate their principal component scores. The ability to classify the calculated principal component scores of [[Image:PCA_Subjectbootstrap.png]] into disease groups was tested by calculation the partial probabilities for membership of each [[Image:PCA_Subjectbootstrap.png]] in a given diagnostic group according to the formula: | To validate the hypothesis that PCA can correctly classify disease groups based on cerebellar shape characteristics alone, we performed a bootstrap validation, leaving out each subject ([[Image:PCA_Subjectbootstrap.png]]) one at a time. The PCA space was defined without the anatomic information of [[Image:PCA_Subjectbootstrap.png]]. The relative regional volumes of [[Image:PCA_Subjectbootstrap.png]] were regressed against the anatomic scores of all subjects minus [[Image:PCA_Subjectbootstrap.png]] within the PCA space to calculate their principal component scores. The ability to classify the calculated principal component scores of [[Image:PCA_Subjectbootstrap.png]] into disease groups was tested by calculation the partial probabilities for membership of each [[Image:PCA_Subjectbootstrap.png]] in a given diagnostic group according to the formula: | ||
| − | + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | |
| − | Equation 1: [[Image:PCA_Equation_1.png]] Equation 2: [[Image:PCA_Equation_2.png]] Equation 3: [[Image:PCA_Equation_3.png]] | + | {| |
| + | |- | ||
| + | |Equation 1: [[Image:PCA_Equation_1.png]] Equation 2: [[Image:PCA_Equation_2.png]] Equation 3: [[Image:PCA_Equation_3.png]] | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| Line 41: | Line 51: | ||
{{h3|Results}} | {{h3|Results}} | ||
| − | [[Image:PCA_Figure_1.png ]] | + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> |
| − | + | {| | |
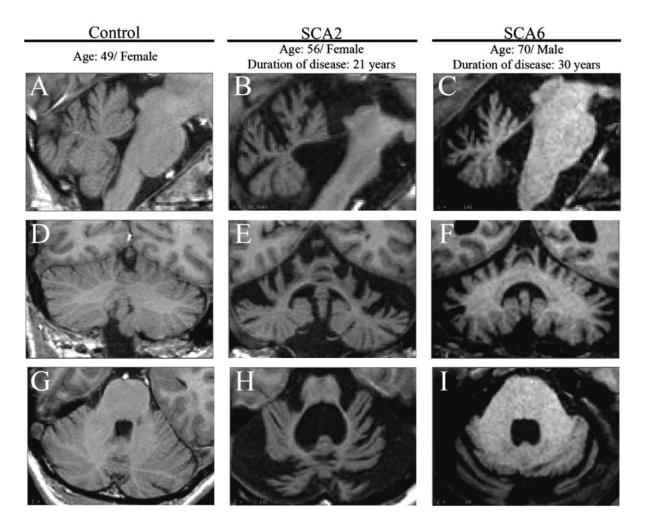
| − | '''Figure 2''': SCAs show disease-specific pattern of regional cerebellar atrophy. Sagittal (a–c), coronal (d–f), and axial (g–i) views of the cerebellum. As compared to control (d) and SCA6 (f), SCA2 (e) shows significant atrophy of the corpus medullare (central white matters of the cerebellum and the deep cerebellar nuclei). Furthermore, as compared to SCA6 (f), SCA2 shows relative sparing of the posterior-inferior regions of the cerebellum | + | |- |
| + | |align=center|[[Image:PCA_Figure_1.png ]] | ||
| + | |- | ||
| + | |'''Figure 2''': SCAs show disease-specific pattern of regional cerebellar atrophy. Sagittal (a–c), coronal (d–f), and axial (g–i) views of the cerebellum. As compared to control (d) and SCA6 (f), SCA2 (e) shows significant atrophy of the corpus medullare (central white matters of the cerebellum and the deep cerebellar nuclei). Furthermore, as compared to SCA6 (f), SCA2 shows relative sparing of the posterior-inferior regions of the cerebellum | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| Line 50: | Line 66: | ||
In post-hoc analysis, the principal component scores of one patient with SCA3, on patient with EA2 and three patients with ICLOCA were calculated as per the method described as the bootstrap validation. The SCA3 patient showed the highest partial probability for membership in SCA2, while the EA2 patient was closer to SCA6. All three patients with ILOCA showed the highest probability of resembling SCA6 phenotype. | In post-hoc analysis, the principal component scores of one patient with SCA3, on patient with EA2 and three patients with ICLOCA were calculated as per the method described as the bootstrap validation. The SCA3 patient showed the highest partial probability for membership in SCA2, while the EA2 patient was closer to SCA6. All three patients with ILOCA showed the highest probability of resembling SCA6 phenotype. | ||
| − | '''Bootstrap validation confirms that anatomic information can predict diagnosis''' | + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> |
| − | + | {| | |
| − | [[Image:PCA_Table_3.png]] | + | |- |
| − | + | |align=center|'''Bootstrap validation confirms that anatomic information can predict diagnosis''' | |
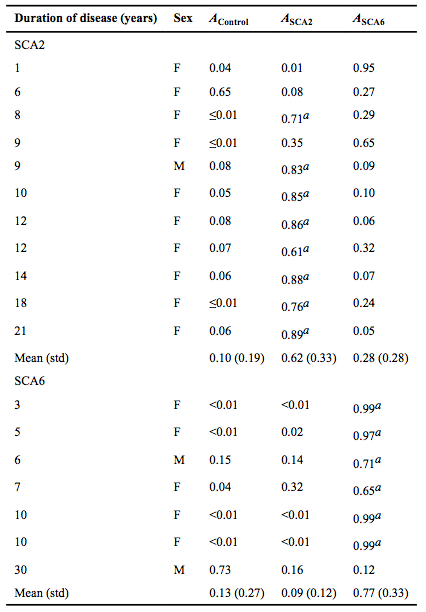
| − | '''Table 1''': A bootstrap validation was performed by leaving out each subject ([[Image:PCA_Subjectbootstrap.png]]) one at a time. PCA was then performed on all subjects minus [[Image:PCA_Subjectbootstrap.png]]. The principal component scores of [[Image:PCA_Subjectbootstrap.png]] were computed by regressing the relative regional volumes of [[Image:PCA_Subjectbootstrap.png]] against the anatomic scores of all subjects minus [[Image:PCA_Subjectbootstrap.png]]. Partial probabilities for membership of [[Image:PCA_Subjectbootstrap.png]] in a given diagnostic group were determined based on the computed principal component scores. [[Image:PCA_Ai.png ]] is the partial probability of membership for diagnostic group [[Image:PCA_i.png]] (control, SCA2, or SCA6) a Indicates subjects whose predicted diagnosis matched their actual diagnosis (partial probability was equal to or greater than the threshold determined by the ROC curve; SCA2=0.52 and SCA6=0.52). Based on the cerebellar shape characteristics alone, 72.7% (8/11) of SCA2 subjects were correctly classified as SCA2, while 85.7% (6/7) of SCA6 subjects were correctly assigned as SCA6'' | + | |- |
| + | |align=center|[[Image:PCA_Table_3.png]] | ||
| + | |- | ||
| + | |'''Table 1''': A bootstrap validation was performed by leaving out each subject ([[Image:PCA_Subjectbootstrap.png]]) one at a time. PCA was then performed on all subjects minus [[Image:PCA_Subjectbootstrap.png]]. The principal component scores of [[Image:PCA_Subjectbootstrap.png]] were computed by regressing the relative regional volumes of [[Image:PCA_Subjectbootstrap.png]] against the anatomic scores of all subjects minus [[Image:PCA_Subjectbootstrap.png]]. Partial probabilities for membership of [[Image:PCA_Subjectbootstrap.png]] in a given diagnostic group were determined based on the computed principal component scores. [[Image:PCA_Ai.png ]] is the partial probability of membership for diagnostic group [[Image:PCA_i.png]] (control, SCA2, or SCA6) a Indicates subjects whose predicted diagnosis matched their actual diagnosis (partial probability was equal to or greater than the threshold determined by the ROC curve; SCA2=0.52 and SCA6=0.52). Based on the cerebellar shape characteristics alone, 72.7% (8/11) of SCA2 subjects were correctly classified as SCA2, while 85.7% (6/7) of SCA6 subjects were correctly assigned as SCA6'' | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| Line 74: | Line 96: | ||
{{h3|Publications}} | {{h3|Publications}} | ||
| − | + | {{iacl-pub|author=B.C. Jung, S.I. Choi, A.X. Du, J.L. Cuzzocreo, Z.Z. Geng, H.S. ying, S.L. Perlman, A.W. Toga, J.L. Prince, and S.H. Ying|title=Principal Component Analysis of Cerebellar Shape on MRI Seperates SCA Types 2 and 6 into Two Archetypal Modes of Degeneration|jrnl= Cerebellum|number=11(4):887-895|when=2012|doi= 10.1007/s12311-011-0334-6}} | |
| − | + | {{iacl-pub|author=B.C. Jung, S.I. Choi, A.X. Du, J.L. Cuzzocreo, H.S. Ying, B.A. Landman, S.L. Perlman, R.W. Baloh, D.S. Zee, A.W. Toga, J.L. Prince, and S.H. Ying|title=MRI shows a region-specific pattern of atrophy in spinocerebellar ataxia type 2|jrnl= Cerebellum|when=2011|doi= 10.1007/s12311-0308-8}} | |
{{h3|References}} | {{h3|References}} | ||
| − | * Schmahmann, JD. "MRI atlas of the human cerebellum, | + | * Schmahmann, JD. '''"MRI atlas of the human cerebellum"''', ''San Diego: Academic Press'', 2000. |
Latest revision as of 00:41, 3 July 2022
Principal Component Analysis of Cerebellar Shape
Brian C. Jung, Soo I. Choi, Annie X. Du, Jennifer L. Cuzzocreo, Zhuo Z. Geng, Howard S. Ying, Susan L. Perlman, Arthur W. Toga, Jerry L. Prince, and Sarah H. Ying
Introduction
Cerebellar ataxias are characterized by poor control of gait, speech, coordination, and eye movements. Anatomically, cerebellar ataxias show progressive atrophy of the cerebellum that is often accompanied by atrophy of the brainstem, cerebral cortex, and other regions. An anatomic biomarker could be an invaluable surrogate for following disease, particularly for patients of more advanced age as confounding medical issues may affect neurologic performance even on standardized ataxia rating scales. In this case we use magnetic resonance (MR) shape characteristics to classify neurodegenerative ataxias into archetypal modes of degeneration. Specifically, we quantify shape based on the relative volumes of the individual lobules that comprise the total cerebellum.
We hypothesize that principal component analysis (PCA) of cerebellar shape characteristics will separate different disease groups into different archetypes based on the differential patterns of cerebellar atrophy. Our central model is that the structure of the cerebellum is related to clinical phenotype and may well be more sensitive to the progression of the disease because structure directly connects to potential underlying mechanisms. We have previously reported disease-specific anatomic differences in a subset of the cerebellar system in spinocerebellar ataxia (SCA) types 2 and 6. Presumably, these region-specific anatomic differences reflect disease-specific pathogenetic mechanisms. In this exploratory paper, we extend our analyses to the entire cerebellum to investigate the possible presence of archetypal modes of cerebellar neurodegeneration.
Methods
We had eleven patients with SCA2 (10 female/1 male) and seven patients with SCA6 (5 female/2 male) were compared against 15 neurologically normal controls (14/1 male). All participants completed a medical questionnaire including items evaluating course of illness, past medical history, and a review of systems. The duration of the disease was defined from the first self-reported symptom of ataxia. Subjects were scanned on either a 1.5 GE Signa scanner with 3D-SPGR sequence or a 3.0T Philips Integra MR scanner. Available scans from a given scanning session were co-registered and averaged using the Brain Imaging Software Toolbox in order to improve signal-to-noise ratio.
Regions of interest were manually delineated using Display software by two raters who were blinded to the diagnosis of participants. Inter-rater reliability of the volumetric measurements was determined by having each rater analyze four subjects for determination of intraclass correlation coefficients. Regions of interest were visually identified based on Schmahmann’s cerebellar atlas. The total cerebellar volume was defined to include the cerebellar cortex, arbor vitae corpus medullare, and deep cerebellar nuclei. In the presence of cerebellar atrophy, the volumetric measurements excluded the cerebrospinal fluid space between the lobules of the cerebellum.
Our study revolved around Principal Component Analysis (PCA). PCA is a mathematical procedure that uses an orthogonal transformation to convert a set of possibly correlated variables into a smaller or equal number of uncorrelated variables called principal components. The first principal component accounts for as much of the variability in the data as possible, and each succeeding component accounts for as much of the remaining variability as possible. PCA was performed on the relative regional volumes and inter-diagnosis differences in principal components were assessed using Hotelling’s T-squared distribution test.
To validate the hypothesis that PCA can correctly classify disease groups based on cerebellar shape characteristics alone, we performed a bootstrap validation, leaving out each subject (![]() ) one at a time. The PCA space was defined without the anatomic information of
) one at a time. The PCA space was defined without the anatomic information of ![]() . The relative regional volumes of
. The relative regional volumes of ![]() were regressed against the anatomic scores of all subjects minus
were regressed against the anatomic scores of all subjects minus ![]() within the PCA space to calculate their principal component scores. The ability to classify the calculated principal component scores of
within the PCA space to calculate their principal component scores. The ability to classify the calculated principal component scores of ![]() into disease groups was tested by calculation the partial probabilities for membership of each
into disease groups was tested by calculation the partial probabilities for membership of each ![]() in a given diagnostic group according to the formula:
in a given diagnostic group according to the formula:
Where ![]() is the partial probability of the membership for diagnostic group
is the partial probability of the membership for diagnostic group ![]() (control, SCA2 or SCA6).
(control, SCA2 or SCA6). ![]() is the set of elements projected onto the vector between the center of mass of the diagnostic group in the PCA space and the principal component scores of subjects
is the set of elements projected onto the vector between the center of mass of the diagnostic group in the PCA space and the principal component scores of subjects ![]() in diagnostic group
in diagnostic group ![]() (table 1). For each subject, the magnitude of the vector between the subject’s principal component scores and the center of mass of each diagnostic group (
(table 1). For each subject, the magnitude of the vector between the subject’s principal component scores and the center of mass of each diagnostic group (![]() ) was computed
) was computed ![]() . Z-score for diagnostic group
. Z-score for diagnostic group ![]() (
(![]() ) for each subject was computed for each disease group based on the magnitude of the vectors between the principal component scores of subjects
) for each subject was computed for each disease group based on the magnitude of the vectors between the principal component scores of subjects ![]() in diagnostic group
in diagnostic group ![]() and the center of mass of the diagnostic group
and the center of mass of the diagnostic group ![]() . Probability (
. Probability (![]() ) for diagnostic group
) for diagnostic group ![]() was defined as 1-normal cumulative distribution of Zb and partial probability (
was defined as 1-normal cumulative distribution of Zb and partial probability (![]() ) was calculated as the ratio of
) was calculated as the ratio of ![]() /sum
/sum![]() . Reciever operation characteristic (ROC) analysis was performed on partial probabilities (
. Reciever operation characteristic (ROC) analysis was performed on partial probabilities (![]() ) to determine the power of the PCA space to discriminate disease groups.
) to determine the power of the PCA space to discriminate disease groups.
Results
The relative volume of corpus medullare in SCA2 subjects differed from the controls, while Crus I trended towards differing. SCA6 did not differ from the controls. SCA2 differed from SCA6 in the relative volume of corpus medullare and nodulus. SCA2 and SCA6 both differed from the controls within the first three principal components. SCA2 also differed from SCA6. The partial probabilities (the likelihood of resembling controls, SCA2, or SCA6 based on cerebellar shape characteristics) for each subject was calculated via a bootstrap validation (Table 3). ROC analysis of the partial probabilities showed that for SCA2, sensitivity and specificity were optimized (α =0.05) by a cut-off point at ASCA2=0.52, with accuracy of 72.7% and an area under the curve (AUC) of 0.826 (p<0.001). For SCA6, sensitivity and specificity were optimized by a cut-off point at ASCA6=0.52, with accuracy of 85.7% and an AUC of 0.852 (p<0.001).
In post-hoc analysis, the principal component scores of one patient with SCA3, on patient with EA2 and three patients with ICLOCA were calculated as per the method described as the bootstrap validation. The SCA3 patient showed the highest partial probability for membership in SCA2, while the EA2 patient was closer to SCA6. All three patients with ILOCA showed the highest probability of resembling SCA6 phenotype.
This is the first study to investigate whether shape characteristics of the cerebellum can predict the diagnosis in unsupervised fashion. Even with the exclusion of the volumetric measurements for pons and the cerebral cortex, which are disproportionately atrophied in SCA2 but spared in SCA6, PCA was able to separate the three groups based solely on cerebellar measurements. The bootstrap validation of SCA2 and SCA6 patients has confirmed that these two disease groups, which have complementary clinical phenotypes, also differ in anatomic characteristics. SCA2 shows slowing of saccades but sparing of smooth pursuit. This pattern of clinical phenotypes are secondary to differential disease specific pattern of atrophy of the flocculus but relative sparing of the pons.
The OCA of cerebellar shape characteristics showed that ILOCA subjects are closer to an unidentified point of convergence in the PCA space that is unique to sporadic forms of ataxia. Alternatively, it may be possible that our ILOCA subjects and SCA6 subjects share similar pathogenetic mechanisms. The patients with sporadic forms of ataxia show a heterogeneous clinical phenotype without identifiable genetic mutation. While our paticular set of ILOCA subjects did not vary widely in anatomic phenotype, a study of a larger sample of ILOCA subjects may separate patients into different archetypal modes of degeneration. If the similarity in the cerebellum could aid in diagnosis, prognosis, and disease staging for sporadic forms of ataxia. This is important for sporadic ataxia, which is the most common form of cerebellar ataxia, and the pathophysiology is less well understood.
Conclusion
PCA of manually parcellated cerebellar lobules on structural MRI produces a shape index that is able to separate controls, SCA2, and SCA6 on cerebellar measurements alone. This relationship is specific to the cerebellum; pons and cerebral cortex wee excluded from the PCA analysis. Lobule-specific characterization of the cerebellar neuro-degeneration could distill complex anatomic variability into clinically meaningful patterns. This could aid in elucidating the mechanisms underlaying the pathogenesis of the disease specifically in sporadic forms of ataxia where the pathophysiology is less well understood.
Acknowledgements
This work was supported by the Arnold-Chiari Foundation, the Robin Zee Fund, the Dana Foundation Program for Brain and Immuno-Imaging, the Research to Prevent Blindness Core Grant, and the National Institutes of Health (grant numbers 1K23EY015802, 5T32DC00023, 5T32MH019950, 5T32GM007057, R01 EY01849, 1R01NS056307, R01NS054255, 5RC1NS068897, 5R01EY019347, and 5R21NS059830). We would also like to thank Mimi Lee and Elizabeth Murray for their technical assistance.
Publications
- B.C. Jung, S.I. Choi, A.X. Du, J.L. Cuzzocreo, Z.Z. Geng, H.S. ying, S.L. Perlman, A.W. Toga, J.L. Prince, and S.H. Ying, "Principal Component Analysis of Cerebellar Shape on MRI Seperates SCA Types 2 and 6 into Two Archetypal Modes of Degeneration", Cerebellum, 11(4):887-895, 2012.

- B.C. Jung, S.I. Choi, A.X. Du, J.L. Cuzzocreo, H.S. Ying, B.A. Landman, S.L. Perlman, R.W. Baloh, D.S. Zee, A.W. Toga, J.L. Prince, and S.H. Ying, "MRI shows a region-specific pattern of atrophy in spinocerebellar ataxia type 2", Cerebellum, 2011.

References
- Schmahmann, JD. "MRI atlas of the human cerebellum", San Diego: Academic Press, 2000.