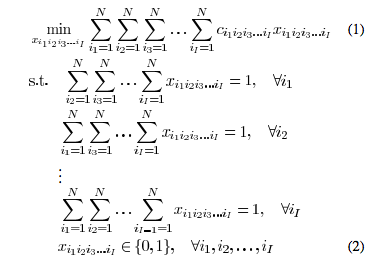Difference between revisions of "Prostate Brachytherapy Intraoperative Seed Reconstruction"
(New page: <meta name="title" content="REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction"/> {{h2|REDMAPS: Reduced-Dimensionality Matching for Prostate Brachyther...) |
|||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | <meta name="title" content="REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction"/> | + | <!-- <meta name="title" content="REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction"/> --> |
{{h2|REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction}} | {{h2|REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction}} | ||
{{TOCright}} | {{TOCright}} | ||
| Line 21: | Line 21: | ||
{{h3|Previous Methods}} | {{h3|Previous Methods}} | ||
| + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | ||
{|align=center | {|align=center | ||
| − | |||
| − | |||
| | | | ||
{| | {| | ||
| Line 33: | Line 32: | ||
|} | |} | ||
|} | |} | ||
| − | + | </div> | |
| Line 43: | Line 42: | ||
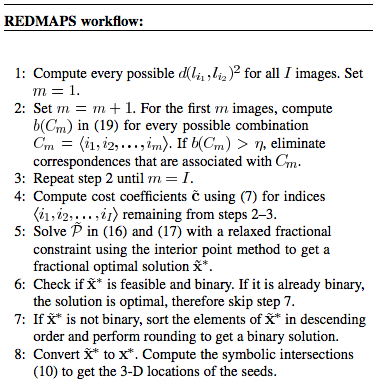
For a general care when at least three [[Image:I-3.png]] images are used and all the N seeds are segmented with their 2-D image coordinates in every image, the seed reconstruction problem can be formulated as a 3-D assignment problem (3DAP) in the following way: | For a general care when at least three [[Image:I-3.png]] images are used and all the N seeds are segmented with their 2-D image coordinates in every image, the seed reconstruction problem can be formulated as a 3-D assignment problem (3DAP) in the following way: | ||
| + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | ||
{|align=center | {|align=center | ||
| − | |||
| − | |||
| | | | ||
{| | {| | ||
| Line 53: | Line 51: | ||
|} | |} | ||
|} | |} | ||
| − | + | </div> | |
where equation [[Image:Xi1i3i3.png]] is equal to one when the match [[Image:-i1i2i3-.png]] is chosen and is zero otherwise, and [[Image:Ci1i2i3.png ]] are the costs associated with the matches. This integer programming problem requires the matching of each and every seed in any one image with unique seeds in each of the other images in such a way that the additive cost associated with all matched seeds is minimized. The cost associated with any [[Image:ItalicizedI.png]] paired seeds can be defined in several ways, and should be large if the paired seeds are not actually associated with a single seed in 3-D. | where equation [[Image:Xi1i3i3.png]] is equal to one when the match [[Image:-i1i2i3-.png]] is chosen and is zero otherwise, and [[Image:Ci1i2i3.png ]] are the costs associated with the matches. This integer programming problem requires the matching of each and every seed in any one image with unique seeds in each of the other images in such a way that the additive cost associated with all matched seeds is minimized. The cost associated with any [[Image:ItalicizedI.png]] paired seeds can be defined in several ways, and should be large if the paired seeds are not actually associated with a single seed in 3-D. | ||
| Line 60: | Line 58: | ||
Dimensionality reduction is possible since the optimal solution has approximately zero cost when the poses of the acquired images are known to be within a small error. REDMAPS is also formulated to address the “hidden seed problem” in which seeds overlap on one or more observed images. REDMAPS uses a pruning algorithm to avoid unnecessary computation of cost metrics and the reduced problem is solved using linear programing. The following describes the overall REDMAPS workflow. It takes 2-D seed coordinates and the image poses as inputs, and outputs the 3-D locations of the reconstructed seeds along with the resolved seed correspondences. | Dimensionality reduction is possible since the optimal solution has approximately zero cost when the poses of the acquired images are known to be within a small error. REDMAPS is also formulated to address the “hidden seed problem” in which seeds overlap on one or more observed images. REDMAPS uses a pruning algorithm to avoid unnecessary computation of cost metrics and the reduced problem is solved using linear programing. The following describes the overall REDMAPS workflow. It takes 2-D seed coordinates and the image poses as inputs, and outputs the 3-D locations of the reconstructed seeds along with the resolved seed correspondences. | ||
| + | |||
| + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | ||
{|align=center | {|align=center | ||
| − | |||
| − | |||
| | | | ||
{| | {| | ||
| Line 70: | Line 68: | ||
|} | |} | ||
|} | |} | ||
| − | + | </div> | |
| Line 83: | Line 81: | ||
REDMAPS also managed to handle seed segmentation errors better than X-MARSHAL and had a smaller reconstruction error with a near perfect matching rate (>99%). For calibration errors REDMAPS also managed to have a near perfect matching rate (>99%) and performed better than X-MARSHAL. Rotation and translation pose errors do bring down the overall matching rate to >97.5% accuracy with up to 2 degree rotation error and 5mm translation error. But within the pose error range of contemporary tracking systems such as FTRAC [Med. Phys. 2005] achieve, it can perform almost perfectly and better than X-MARSHAL. | REDMAPS also managed to handle seed segmentation errors better than X-MARSHAL and had a smaller reconstruction error with a near perfect matching rate (>99%). For calibration errors REDMAPS also managed to have a near perfect matching rate (>99%) and performed better than X-MARSHAL. Rotation and translation pose errors do bring down the overall matching rate to >97.5% accuracy with up to 2 degree rotation error and 5mm translation error. But within the pose error range of contemporary tracking systems such as FTRAC [Med. Phys. 2005] achieve, it can perform almost perfectly and better than X-MARSHAL. | ||
| + | |||
| + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | ||
{|align=center | {|align=center | ||
| − | |||
| − | |||
| | | | ||
{| | {| | ||
| Line 95: | Line 93: | ||
|} | |} | ||
|} | |} | ||
| − | + | </div> | |
{{h3|ACP-REDMAPS}} | {{h3|ACP-REDMAPS}} | ||
| Line 103: | Line 101: | ||
We have also demonstrated a seed reconstruction scenario without using a tracking device. Even when no tracking device was used and the initial pose was roughly estimated under a simplified geometry, ACP-REDMAPS was still able to recover image poses and achieve near perfect seed detection rate. Although we relied on the angular mark readings on the C-arm for the initial poses, the results on clinical data sets showed that APC-REDMAPS can accommodate very rough initial guess on the image poses and has a potential for a trackless seed reconstruction strategy can also be adopted in order to guarantee more accurate initialization. Overall, ACP-REDMAPS can greatly improve the seed detection rate even from the already good performance of REDMAPS without requiring a significant amount of additional time. This approach not only allows a more generous choice of tracking methods but also opens a possibility to completely remove the use of any external trackers. | We have also demonstrated a seed reconstruction scenario without using a tracking device. Even when no tracking device was used and the initial pose was roughly estimated under a simplified geometry, ACP-REDMAPS was still able to recover image poses and achieve near perfect seed detection rate. Although we relied on the angular mark readings on the C-arm for the initial poses, the results on clinical data sets showed that APC-REDMAPS can accommodate very rough initial guess on the image poses and has a potential for a trackless seed reconstruction strategy can also be adopted in order to guarantee more accurate initialization. Overall, ACP-REDMAPS can greatly improve the seed detection rate even from the already good performance of REDMAPS without requiring a significant amount of additional time. This approach not only allows a more generous choice of tracking methods but also opens a possibility to completely remove the use of any external trackers. | ||
| + | <div style="background: white; border: 2px solid rgb(150, 150, 150); padding: 2px; text-align: justify;"> | ||
{|align=center | {|align=center | ||
| − | |||
| − | |||
| | | | ||
{| | {| | ||
| Line 115: | Line 112: | ||
|} | |} | ||
|} | |} | ||
| − | + | </div> | |
Latest revision as of 00:45, 3 July 2022
REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction
Junghoon Lee, Christian Labat, Ameet K. Jain, Danny Y. Song, Everette Clif Burdette, Gabor Fichtinger, and Jerry L. Prince
Introduction
Carcinoma of the prostate s one of the most prevalent and fatal cancers for North American men, making up 25% of all cases and 9% of deaths for men. In the contemporary prostate brachytherapy procedure, and implant plan is made either preoperatively or intraoperatively based on transrectal ultrasound (TRUS) imaging. One of the greatest challenges of the current TRUS-guided implant method is that it is difficult to visualize the implanted seeds on TRUS. As a consequence, seed positioning variations cannot be identified intraoperatively during the procedure. Sometimes an additional seed implant session or supplemental external beam radiation is required. Existing seed reconstruction methods have at least one of the fundamental limitations:
- They are sensitive to pose errors of the imaging device
- They cannot resolve the hidden seed problem
- They require a large number of images, large image acquisition angles, and a long time to compute a converging solution
- They require constrained motion of the imaging device, e.g., isocentric circular trajectory
The approach described in this paper addresses all of these problems and was designed to be robust to these limitations and to work on any type of X-ray imaging system. This is critical because all four limitations must be solved in order for the method to be suitable for standard clinical use. We present a seed-matching algorithm that is able to find the 3-D seed locations from multiple X-ray images in the presence of hidden seeds and pose errors of the imaging device in polynomial time. We named our algorithm REDMAPS for REduced-Dimensionality Matching Algorithm for Prostate brachytherapy Seed reconstruction. REDMAPS adopts a dimensionality reduction approach to efficiently solve the NP-hard combinatorial optimization problem. The key insight to dimensionality reduction is the practical observation that the optimal solution of our problem has near-zero const if the pose of the imaging device is known to within a small error. We also apply a pruning algorithm for efficient cost computation that allows us to significantly reduce the total computation time. The reduced problem is efficiently solved using linear programming in polynomial time.
REDMAPS was designed to work on conventional mobile C-arms, which are often available to most brachytherapy practitioners. Importantly REDMAPS is able to provide implant reconstruction at the end of the procedure for the purpose of exit dosimetry quality assurance when there is the largest number of seeds and correspondingly higher rates of overlaps. Finally, although REDMAPS assumes that the image acquisition poses are known, it was designed to be robust to the level of a realistic pose errors that is commonly observed in mobile C-arm applications.
Previous Methods
Before REDMAPS, we proposed a tomosynthesis-based prostate brachytherapy seed reconstruction method. The attractive feature of the proposed method is that it can automatically recover all the seeds, including overlapping or clustered seeds, without involving explicit identification of the 2-D coordinates of the seeds in the fluoroscopy images. Gaussian blurring combined with distance map enables the algorithm to reconstruct seeds under realistic calibration and pose estimation errors of the C-arm. In addition, a C-arm auto-focus process enables the algorithm to utilize out of focus images after automatic adjustment of the C-arm camera parameters. This proposed algorithm required three to four images to detect the implanted seeds with a detection accuracy of (greater than or equal to) 97.9% when the C-arm is accurately estimated with angle separation (greater than or equal to) 10 degrees. In case the image acquisition angles are very small, it is natural for the algorithm to have difficulty in detecting all true seeds correctly. However, simulations with varying angular separation show that the algorithm was robust to angle separation up to five degree cone angle centered on the AP-axis. Overall, the algorithm performed well, but is not as competent as our current REDMAPS algorithm.
What is REDMAPS
For a general care when at least three ![]() images are used and all the N seeds are segmented with their 2-D image coordinates in every image, the seed reconstruction problem can be formulated as a 3-D assignment problem (3DAP) in the following way:
images are used and all the N seeds are segmented with their 2-D image coordinates in every image, the seed reconstruction problem can be formulated as a 3-D assignment problem (3DAP) in the following way:
where equation ![]() is equal to one when the match
is equal to one when the match ![]() is chosen and is zero otherwise, and
is chosen and is zero otherwise, and ![]() are the costs associated with the matches. This integer programming problem requires the matching of each and every seed in any one image with unique seeds in each of the other images in such a way that the additive cost associated with all matched seeds is minimized. The cost associated with any
are the costs associated with the matches. This integer programming problem requires the matching of each and every seed in any one image with unique seeds in each of the other images in such a way that the additive cost associated with all matched seeds is minimized. The cost associated with any ![]() paired seeds can be defined in several ways, and should be large if the paired seeds are not actually associated with a single seed in 3-D.
When
paired seeds can be defined in several ways, and should be large if the paired seeds are not actually associated with a single seed in 3-D.
When ![]() , 3DAP given by (1) and (2) is NP-hard and is generally impractical to solve exactly except when there is a very small number of seeds. Thus, approximate solutions are predominant in the literature. Also, the problem statement does not account for the presence of hidden seeds, which becomes inevitable as the number of seeds rises to a therapeutically relevant level. Although solutions to the hidden seed problem exist, they are either computationally intensive or ultimately require manual intervention. In the following sections, we extend the problem statement to include hidden seeds and then find the solution by using dimensionality reduction. The algorithm turns out to be computationally feasible in clinically relevant scenarios.
, 3DAP given by (1) and (2) is NP-hard and is generally impractical to solve exactly except when there is a very small number of seeds. Thus, approximate solutions are predominant in the literature. Also, the problem statement does not account for the presence of hidden seeds, which becomes inevitable as the number of seeds rises to a therapeutically relevant level. Although solutions to the hidden seed problem exist, they are either computationally intensive or ultimately require manual intervention. In the following sections, we extend the problem statement to include hidden seeds and then find the solution by using dimensionality reduction. The algorithm turns out to be computationally feasible in clinically relevant scenarios.
Dimensionality reduction is possible since the optimal solution has approximately zero cost when the poses of the acquired images are known to be within a small error. REDMAPS is also formulated to address the “hidden seed problem” in which seeds overlap on one or more observed images. REDMAPS uses a pruning algorithm to avoid unnecessary computation of cost metrics and the reduced problem is solved using linear programing. The following describes the overall REDMAPS workflow. It takes 2-D seed coordinates and the image poses as inputs, and outputs the 3-D locations of the reconstructed seeds along with the resolved seed correspondences.
Results of REDMAPS Testing
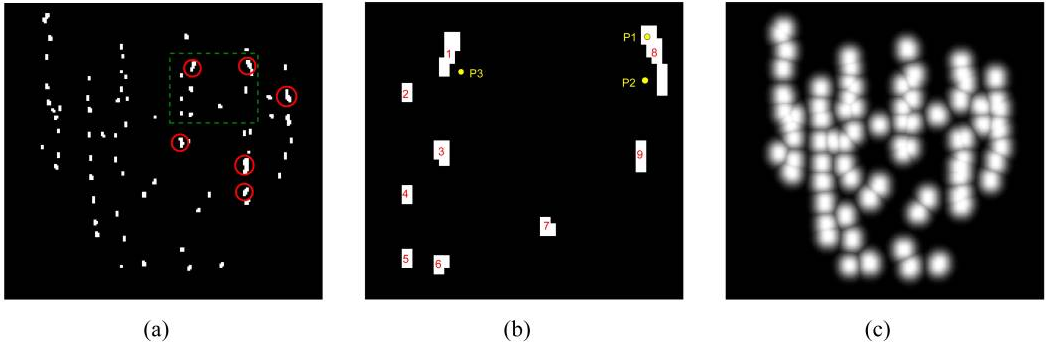
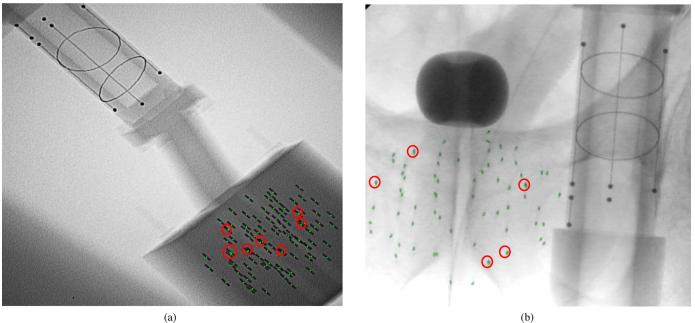
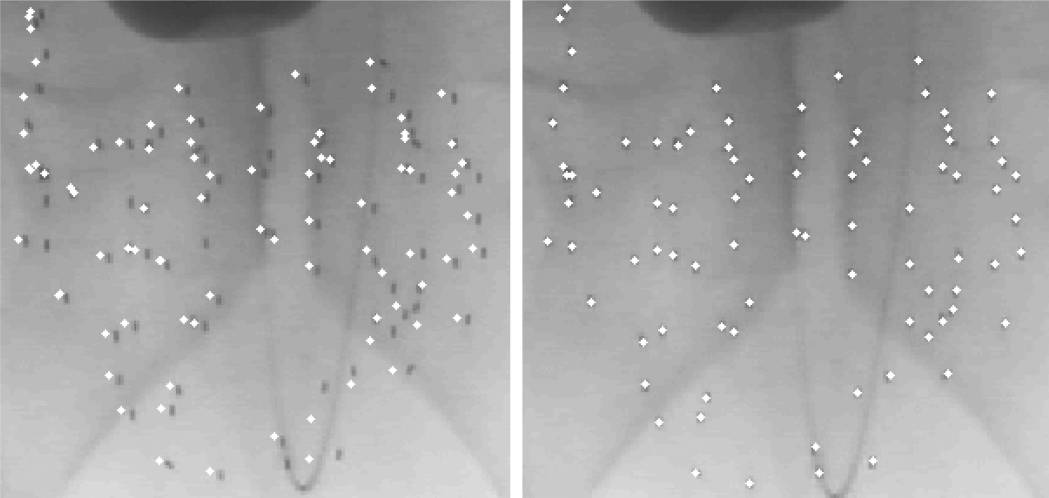
In our phantom experiments and clinical study, fluoroscopy images were acquired using mobile C-arms with an X-ray image intensifier (XRII). The pose of the C-arms was estimated by using a fluoroscope tracking fiducial (FTRAC) [Med Phys. 2005]. Once an image was acquired, it was first preprocessed for geometric distortion correction using precomputed distortion parameters in the calibration step. The FTRAC fiducial and the seeds were segmented from distortion-corrected images using algorithms developed by Kuo et. al. [SPIE 2010]. The estimated C-arm camera parameters and the segmented seeds (with their 2-D image coordinates) are the input to REDMAPS.
Assuming a set of clinically realistic C-arm imaging parameters and seed configuration, we created synthetic projection images. In order to evaluate the robustness of REDMAPS to various errors, we added random errors that are uniformly distributed to the known parameters. All reconstructions were computed first by REDMAPS, and then by the extended-MARSHAL (X-MARSHAL) algorithm [IEEE 2006] which is a fast and reliable seed-matching algorithm for performance comparison.
Seed density increases lead to more complicated matching problems. For our tests we used a total of 1400 reconstructions (7 seed densities x 10 datasets x 4C3 combinations x 5 image acquisition angles) using three images with known poses. Figure 4(a) show the seed matching rate as a function of seed density. Both X-MARSHAL and REDMAPS almost perfectly at resolving the correspondences, but REDMAPS performed slightly better as the seed density increased. We also studied the effectiveness of REDMAPS at various image acquisition angles (as in a real clinical environment the C-arm angles would be limited due to patient position) with angles varying from 5 degrees to 25 degrees. While the narrower the angle became the lower the matching rate was, the seed matching remains almost perfect (>99%) and the reconstruction errors are very small (i.e. less than 0.5mm). While both algorithms performed extremely well, REDMAPS performed better.
REDMAPS also managed to handle seed segmentation errors better than X-MARSHAL and had a smaller reconstruction error with a near perfect matching rate (>99%). For calibration errors REDMAPS also managed to have a near perfect matching rate (>99%) and performed better than X-MARSHAL. Rotation and translation pose errors do bring down the overall matching rate to >97.5% accuracy with up to 2 degree rotation error and 5mm translation error. But within the pose error range of contemporary tracking systems such as FTRAC [Med. Phys. 2005] achieve, it can perform almost perfectly and better than X-MARSHAL.
|
ACP-REDMAPS
In 2011, a new process called Automatic Pose Correction (ACP) was added to the algorithm. ACP is a new pose correction step for accurate reconstructions and localization of the implanted brachytherapy seeds from a small number of X-ray images. By using the reconstructed seeds with revealed seed correspondences as a fiducial, APC iteratively minimizes the RA cost and improves the estimated image poses. As the RA cost becomes smaller at each iteration, the dimensionality reduction threshold in REDMAPS is adaptively reduced to achieve larger dimensionality reduction threshold in REDMAPS is adaptively reduced to achieve larger dimensionality reduction and allow faster computation in the BIP optimization. Both simulations and clinical studies show very promising results where we can achieve almost perfect seed detection rate with very small reconstruction error even when the image pose errors are very large. Especially, as the pose errors are reduced, the resulting solutions become typically binary (on average 99.8% of the solutions were binary in our clinical studies) even under the relaxed fractional constraints, and are therefore globally optimal.
We have also demonstrated a seed reconstruction scenario without using a tracking device. Even when no tracking device was used and the initial pose was roughly estimated under a simplified geometry, ACP-REDMAPS was still able to recover image poses and achieve near perfect seed detection rate. Although we relied on the angular mark readings on the C-arm for the initial poses, the results on clinical data sets showed that APC-REDMAPS can accommodate very rough initial guess on the image poses and has a potential for a trackless seed reconstruction strategy can also be adopted in order to guarantee more accurate initialization. Overall, ACP-REDMAPS can greatly improve the seed detection rate even from the already good performance of REDMAPS without requiring a significant amount of additional time. This approach not only allows a more generous choice of tracking methods but also opens a possibility to completely remove the use of any external trackers.
|
Conclusion
In simulations REDMAPS matched over 99% of the seeds with a reconstruction error of less than 0.5mm on average using three images when there are realistic calibration and pose errors. Further simulations showed that REDMAPS is robust to significantly large errors introduced to seed segmentation, calibration, and pose estimation. REDMAPS was validated on five phantom and 21 clinical datasets and it localized the seeds with the overall seed matching rate over 99% and reconstruction error less than 1mm using only three images. In comparison with X-MARSHAL [IEEE 2006], a fast and reliable seed matching algorithm that is able to recover hidden seeds, REDMAPS was equivalently fast (in the order of seconds) while being more robust to pose errors.
In summary, REDMAPS was found to be sufficiently accurate, robust, and computationally efficient to enable intraoperative dynamic, optimized brachytherapy with conventional low-end C-arms without altering the clinical workflow, thereby allowing for wide-scale clinical deployment in the community care setting.
Publications
- J. Lee, C. Labat, A.K. Jain, D.Y. Song, E.C. Burdette, G. Fitchinger, and J.L. Prince, "REDMAPS: Reduced-Dimensionality Matching for Prostate Brachytherapy Seed Reconstruction", IEEE Transactions on Medical Imaging, 30(1): 38-51, 2011.
- J. Lee, X. Liu, A.K. Jain, D.Y. Song, E.C. Burdette, J.L. Prince, and G. Fichtinger, "Prostate brachytherapy seed reconstruction with Gaussian blurring and optimal coverage cost", IEEE Transactions on Medical Imaging, 28(12):1955-1968, 2009. (PubMed)
- J. Lee, N. Kuo, A. Deguet, E. Dehghan, D.Y. Song, E.C. Burdette, and J.L. Prince, "Intraoperative 3-D Reconstruction of Prostate Brachytherapy Implants with Automatic Pose Correction", Physics in Medicine and Biology, 56(15):5011-5027, 2012. (PubMed)
References
- A. K. Jain, T. Mustafa, Y. Zhou, C. Burdette, G. S. Chirikjian, and G. Fichtinger, “FTRAC—A robust fluoroscope tracking fiducial,” Med. Phys., vol. 32, pp. 3185–3198, 2005
- N. Kuo, J. Lee, A. Deguet, D. Y. Song, E. C. Burdette, and J. L. Prince, “Automatic segmentation of seeds and fluoroscope tracking (FTRAC) fiducial in prostate brachytherapy X-ray images,” in Proc. SPIE Med. Imag., 2010, vol. 7625, pp. 76 252T1–76 252T9.
- R. Kon, A. Jain, and G. Fichtinger, “Hidden seed reconstruction from C-arm images in brachytherapy,” in IEEE Int. Symp. Biomed. Imag., Apr. 2006, pp. 526–529