Magnetic Resonance Image Example Based Contrast Synthesis
<meta name="title" content="Magnetic Resonance Image Example-Based Contrast Synthesis"/>
Magnetic Resonance Image Example-Based Contrast Synthesis
Snehashis Roy, Aaron Carass, and Jerry L Prince
This web page is still being constructed
Introduction
Magnetic resonance (MR) imaging (MRI) is widely used to image the brain. Often times though after the use of an image analysis algorithm the image quality can deteriorate if the image is not acquired according to optimal protocol. There are many practical reasons to disregard protocol such as when making a rapid emergency scan or multi-institutional trials, but it often results with images that are of not the desired tissue contrast or intensity. This paper proposes an image restoration technique called MR image example-based contrast synthesis (MIMECS). The central idea behind MIMECS is to synthesize MR contrasts using patches (small regions of the image). The core idea of MIMECS is that each subject patch can be matched to a combination of a few relevant patches from a dictionary in a nonlocal fashion [43], [45], [46]. It is different from classical histogram matching in the sense that a patch can be thought of as a feature vector of its center voxel, thus including local neighborhood information for that voxel. There are several advantages of MIMECS compared to previous synthesis methods. First, it is a completely automatic preprocessing step that can precede any image processing task. Second, there is no need to use multiple, calibrated pulse sequences in order to estimate underlying tissue parameters. Third, even though MIMECS uses an atlas, there is no need to carry out subject-to-atlas registration, thus avoiding potential errors due to misregistration or missing tissues. Finally, it does not require any segmentation of the subject.
Methods
___________________________________________________________________________________________________________________________________________________________________________
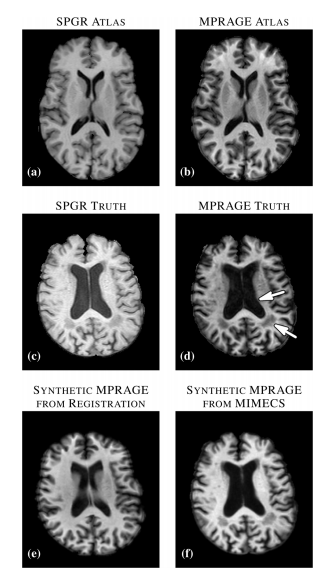
Figure 1: (A) Atlas T1-weighted SPGR and (B) its corresponding T1-weighted MPRAGE. (C) Subject T1-weighted SPGR scan and (D) its T1-weighted MPRAGE image. Atlas SPGR is deformably registered to the subject SPGR. This deformation is applied to the atlas MPRAGE to obtain (E) a synthetic subject MPRAGE. (F) Synthetic MPRAGE image generated by our algorithm, MIMECS.
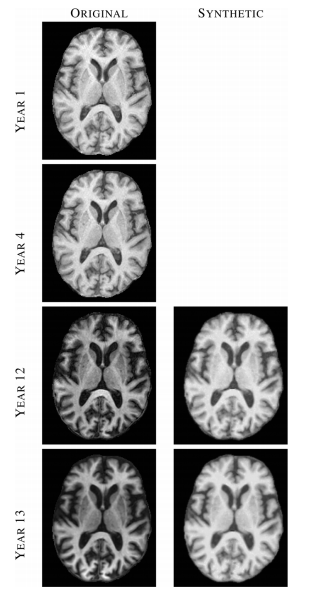
Figure 2: Left column shows images from four (out of 14) time-points of a normal BLSA subject, where each image was acquired approximately one year apart. Right column shows synthetic SPGR images, synthesized using MIMECS applied to the corresponding MPRAGE images in the left column
Figure 1 left, Figure 2 right ___________________________________________________________________________________________________________________________________________________________________________ To understand the work on this paper you must understand the idea of sparse priors. Sparse signal reconstruction maintains that because most signals are sparse in some way it is not necessary to observe the full signal in order to accurately reconstruct it. Sparse signal reconstruction has been applied to many image processing algorithms including denoising, image restoration, and super-resolution. Dictionary-based patch matching methods, popular for natural image restoration, are also used. In our case, we build an overcomplete dictionary using patches from human brain images to both normalize a given contrast and synthesize alternate MR tissue contrasts.
The MIMECS method is based on analysis of image patches. Patches are typically small and centered on the voxel of interest. The voxels within a patch are always ordered in the same way, using a consistent rasterization. For each voxel, a dictionary is found by casting all patches into one higher dimension and searching for “like” patches. A sparse and nonnegative combination of dictionary patches is found to match the subject patch. Then the patches are combined with patches from another dictionary physically matched to the first dictionary. MIMECS also shares strong similarities with nonlocal means methods, some which have been applied in the medical imaging community for noise reduction, super-resolution, and segmentation. An illustration of how the MIMEC Algorithm works is detailed in Fig. 1.
___________________________________________________________________________________________________________________________________________________________________________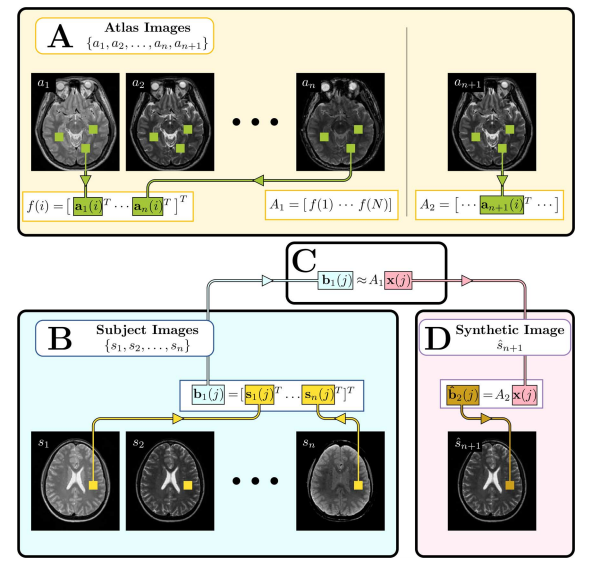
Figure 3 ___________________________________________________________________________________________________________________________________________________________________________ Illustration of the MIMECS algorithm. Region of the image labeled A, shows the construction of the atlases and A2. Region B shows the input subject images converted into patches b1(j). Region C denotes the estimation of the coefficients x(j) which relate b1(j) to the patches of the atlas A1. Finally, region D shows the computation of the synthetic image Sn +1 by using the learned coefficients x(j) to compute the patch b2(j) based on the atlas A2. ___________________________________________________________________________________________________________________________________________________________________________ Results
The most important results from the set of experiments can be summarized as follows. First, we found that features an nose that are present in the subject image but not in the atlas image (defined in the paper) are suppressed to some degree in the reconstruction. This was true in both recovering the same tissue contrast and in recovering different tissue contrast. This suggests that MIMECS can be used for noise suppression and also shows that MIMECS atlas should contain a patch dictionary that is rich enough to encompass the patches that are expected to be found in the source image. Second, the experiments confirmed that sparsity is important when reconstructing patches with significant detail because otherwise the detail will be smoothed out. Third, MIMECS is robust to the specifics of the subject pulse sequence. In paticular, we found that an atlas comprising both an SPGR and MPRAGE can be used to synthesize either an MPRAGE or SPGR image even if the subject image did not precisely match the SPGR or MPRAGE pulse sequence parameters of the atlas. Fourth, we found that MIMECS normalization is comparable or better than histogram matching when normalizing the same tissue contrast and is far more general in that it can recover missing contrasts. Finally, we found we found that errors in MIMECS synthesis do not take on specific anatomical features, but instead resemble a typical noise pattern of MR images.
___________________________________________________________________________________________________________________________________________________________________________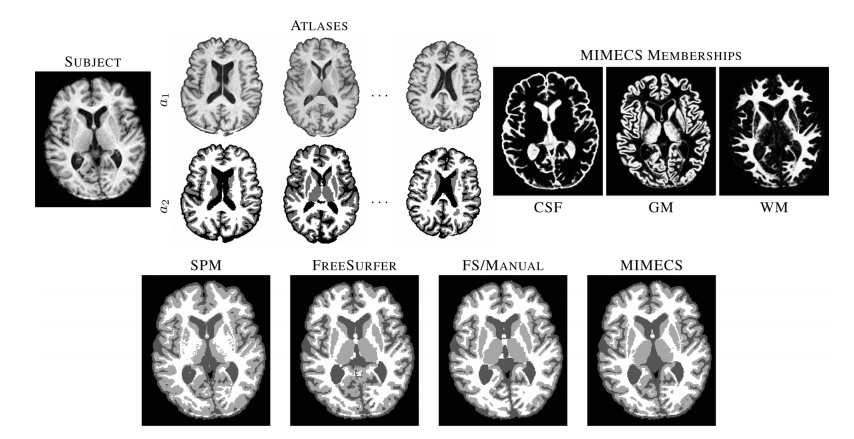
Figure 4 ___________________________________________________________________________________________________________________________________________________________________________ Top row shows a subject T1-w SPGR image, three of the five classification atlases, and fuzzy memberships produced by MIMECS-based tissue classification. Hard segmentations from two leading automatic methods and a manually-corrected method are compared to the hard segmentation of MIMECS. ___________________________________________________________________________________________________________________________________________________________________________
Conclusion
The results demonstrate the potential application of MIMECS, particularly in solving the “missing” or “mis-matched” tissue contrast problem in routine neuroimage processing tasks. The MIMECS framework shares similarities to other patch-based methods, but offers direct synthesis of an alternate tissue contrast. The case study demonstrates the immediate utility of MIMECS for a wide variety of neuroimage processing tasks. MIMECS represents a new class of neuroimage processing methods with a potentially rich future.
Acknowledgements
The authors would like to thank A. Jog for his help in coding the K-d tree and to D.L. Pham for his insightful comments about this manuscript during its preparation. The authors would also like to thank the BLSA participants and neuroimaging staff for their dedication to these studies. Some data used for this study were downloaded from the Biomedical Informatics Research Network (BIRN) Data Repository, supported by grants to the BIRN Coordinating Center (U24-RR019701) Function BIRN (U24-RR021992, Morphometry BIRN (U24-RR021382), and Mouse BIRN (U24-RR021760) Testbeds, funded by the National Center for Research Resources at the National Institutes of Health, USA
References
- S. Roy et al., "Magnetic Resonance Image Example-Based Contrast Synthesis,". IEEE Transactions on Medical Imaging 32, no. 12 (2013). 2348-2363.
- S. Roy et al., "A compressed sensing approach for MR tissue contrast synthesis", IPMI 2011.
- Jerry L Prince, "Atlas-based approach to MR intensity normalization, contrast synthesis, and inhomogeneity correction," Presentation at CSAIL, Cambridge, MA, October 07, 2010.